Genetic insights into Shewanella spp., progenitor of the bla OXA-48-like genes: a large-scale study
- PMID: 40471180
- PMCID: PMC12282331
- DOI: 10.1099/mgen.0.001417
Genetic insights into Shewanella spp., progenitor of the bla OXA-48-like genes: a large-scale study
Abstract
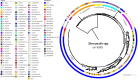
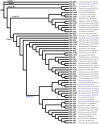
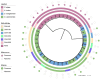
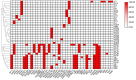
Shewanella spp. played pivotal ecological roles and were reported to be the progenitor of bla OXA-48-like carbapenemase genes. However, it remained unknown which species was the progenitor of different OXA-48 carbapenemase variants. To address this issue, we analysed the largest collection of Shewanella genomes to our knowledge and performed genetic and phenotypic analysis on Shewanella collected from Zhejiang province, China. Our results suggested that bla OXA-48-like was intrinsically carried by a few Shewanella species and different bla OXA-48-like variants were associated with different Shewanella species; for instance, Shewanella baltica was associated with bla OXA-924, and some Shewanella oncorhynchi and Shewanella putrefaciens carried bla OXA-900-like. The bla OXA-48-like genes carried by Shewanella xiamenensis were highly diverse. Comparatively, none of the Shewanella algae genomes carried bla OXA-48-like. Results of phylogenetic analysis supported the notion that OXA-48-like carbapenemase originated from different environmental Shewanella species and was captured by clinical species, particularly Enterobacterales. Different Shewanella species distributed in different niches in Zhejiang province, i.e. S. algae (n=12) and Shewanella indica (n=1) strains were all isolated from clinical settings and S. xiamenensis (n=23) and Shewanella mangrovisoli (n=2) were isolated from both hospital sewage and river water. bla OXA-48-like genes in Shewanella spp. from Zhejiang province were located on the chromosome. To the best of our knowledge, this is the first study investigating the progenitor of different bla OXA-48-like variants with a focus on the Shewanella population. Results in this study highlighted the important role of Shewanella species in the ecosystem, particularly as the major source of the notorious carbapenemase gene, bla OXA-48. Control measures should be implemented to prevent further dissemination of such organisms in the hospital setting and the community.
Keywords: OXA-48-like; Shewanella spp.; genomic characterization; progenitor; transmission.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Unexpected high prevalence of carbapenemase-producing bacteria and widely spread of blaNDM-1-positive Shewanella spp. in retail shrimp.Int J Food Microbiol. 2025 Oct 2;441:111284. doi: 10.1016/j.ijfoodmicro.2025.111284. Epub 2025 May 29. Int J Food Microbiol. 2025. PMID: 40482551
-
Investigation and pathogenetic testing of Shewanella spp. positive diarrhea cases in Beijing, China.Sci Rep. 2025 Aug 19;15(1):30334. doi: 10.1038/s41598-025-15865-1. Sci Rep. 2025. PMID: 40830642 Free PMC article.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Global emergence and transmission dynamics of carbapenemase-producing Citrobacter freundii sequence type 22 high-risk international clone: a retrospective, genomic, epidemiological study.Lancet Microbe. 2025 Jul 16:101149. doi: 10.1016/j.lanmic.2025.101149. Online ahead of print. Lancet Microbe. 2025. PMID: 40683282
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

