Phase-free local ancestry inference mitigates the impact of switch errors on phase-based methods
- PMID: 40471844
- PMCID: PMC12341942
- DOI: 10.1093/g3journal/jkaf122
Phase-free local ancestry inference mitigates the impact of switch errors on phase-based methods
Abstract
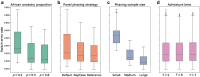
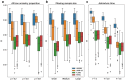
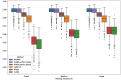
Local ancestry inference is an indispensable component of a variety of analyses in medical and population genetics, from admixture mapping to characterizing demographic history. However, the accuracy of local ancestry inference depends on a number of factors such as phase quality (for phase-based local ancestry inference methods) and time since admixture. Here, we present an empirical analysis of four local ancestry inference methods using simulated individuals of mixed African and European ancestry, examining the impact of variable phase quality and a range of demographic scenarios. We find that regardless of phasing options, calls from local ancestry inference methods that operate on unphased genotypes (phase-free local ancestry inference) have 2.6-4.6% higher Pearson correlation with the ground truth than methods that operate on phased genotypes (phase-based local ancestry inference). Applying the TRACTOR phase correction algorithm led to modest improvements in phase-based local ancestry inference, but despite this, the Pearson correlation of phase-free local ancestry inference remains 2.4-3.8% higher than phase-corrected phase-based approaches (considering the best-performing methods in each category). Further, analyzing perfectly phased data yields accuracies for the phase-based local ancestry inference methods that are only slightly inferior to those of HAPMIX. Phase-free and phase-based local ancestry inference accuracy differences can dramatically impact downstream analyses: estimates of the time since admixture using phase-based local ancestry inference tracts are upwardly biased by ≈10 generations using our highest quality statistically phased data but have virtually no bias using phase-free local ancestry inference calls. This study underscores the strong dependence of phase-based local ancestry inference accuracy on phase quality and highlights the merits of local ancestry inference approaches that analyze unphased genetic data.
Keywords: admixture; ancestry; haplotypes; local ancestry inference; phase.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: A.L.W. holds stock in 23andMe, Inc. and is the owner of HAPI-DNA LLC. S.A. declares no competing interests.
Figures

Update of
-
Phase-free local ancestry inference mitigates the impact of switch errors on phase-based methods.bioRxiv [Preprint]. 2023 Dec 4:2023.12.02.569669. doi: 10.1101/2023.12.02.569669. bioRxiv. 2023. Update in: G3 (Bethesda). 2025 Aug 6;15(8):jkaf122. doi: 10.1093/g3journal/jkaf122. PMID: 38106003 Free PMC article. Updated. Preprint.
References
-
- Atkinson EG, Maihofer AX, Kanai M, Martin AR, Karczewski KJ, Santoro ML, Ulirsch JC, Kamatani Y, Okada Y, Finucane HK, et al. 2021. Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power. Nat Genet. 53(2):195–204. 10.1038/s41588-020-00766-y - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
