Developmental transcriptomics of the Firebrat: Exploring developmental expression patterns and morphology during the embryogenesis of Thermobia domestica
- PMID: 40471973
- PMCID: PMC12140273
- DOI: 10.1371/journal.pone.0324844
Developmental transcriptomics of the Firebrat: Exploring developmental expression patterns and morphology during the embryogenesis of Thermobia domestica
Abstract
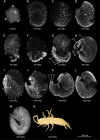
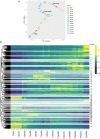
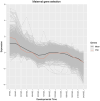
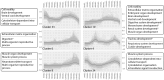
Understanding the development of early-diverging lineages is crucial for inferring evolutionary context in evolutionary developmental biology. Thermobia domestica (the firebrat), a member of Zygentoma, holds particular significance to insect phylogenetics due to its position as a sister group to all winged insects (Pterygota). We explore its development by reporting on the embryonic morphology using DAPI staining at 14 selected timepoints throughout development, which lasts 10 days. At each timepoint, RNA sequencing was conducted to perform a large-scale transcriptomic analysis. Differential gene expression analysis and gene ontology (GO) enrichment studies revealed global expression patterns and linked biological processes to specific developmental stages. Key findings include the identification of major transcriptional turning points during three developmental stages: The maternal to zygotic transition between 16 and 24 hours after egg-laying (hAEL), katatrepsis between 72 and 96 hAEL, and hatching between 216 and 240 hAEL. Additionally, the GO study mapped the timing of biological processes, such as cleavage, blastoderm formation, dorsal closure, and organogenesis, particularly tracheal and muscular development. These insights establish a robust temporal framework for T. domestica embryogenesis and provide a foundation for inferring ancestral and derived developmental traits in insects. The dataset serves as a valuable library for genetic studies and reference for comparative evo-devo studies with other insect species.
Copyright: © 2025 Makkinje et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Mayfly developmental atlas: developmental temporal expression atlas of the mayfly, Ephemera vulgata, reveals short germ-specific hox gene activation.BMC Genomics. 2024 Dec 4;25(1):1177. doi: 10.1186/s12864-024-10934-7. BMC Genomics. 2024. PMID: 39633303 Free PMC article.
-
Selection of Reference Genes for Normalization of Gene Expression in Thermobia domestica (Insecta: Zygentoma: Lepismatidae).Genes (Basel). 2020 Dec 25;12(1):21. doi: 10.3390/genes12010021. Genes (Basel). 2020. PMID: 33375665 Free PMC article.
-
Comparative transcriptomics reveal developmental turning points during embryogenesis of a hemimetabolous insect, the damselfly Ischnura elegans.Sci Rep. 2017 Oct 19;7(1):13547. doi: 10.1038/s41598-017-13176-8. Sci Rep. 2017. PMID: 29051502 Free PMC article.
-
Comparative Transcriptomes and EVO-DEVO Studies Depending on Next Generation Sequencing.Comput Math Methods Med. 2015;2015:896176. doi: 10.1155/2015/896176. Epub 2015 Oct 12. Comput Math Methods Med. 2015. PMID: 26543497 Free PMC article. Review.
-
The developmental hourglass model and recapitulation: An attempt to integrate the two models.J Exp Zool B Mol Dev Evol. 2022 Jan;338(1-2):76-86. doi: 10.1002/jez.b.23027. Epub 2021 Jan 27. J Exp Zool B Mol Dev Evol. 2022. PMID: 33503326 Free PMC article. Review.
References
-
- Grimaldi DA, Engel MS. Evolution of the insects. New York: Cambridge University Press. 2005.
-
- Hall BK. Evolutionary developmental biology (Evo-Devo): Past, present, and future. Evo Edu Outreach. 2012;5(2):184–93. doi: 10.1007/s12052-012-0418-x - DOI
-
- Ohde T, Masumoto M, Yaginuma T, Niimi T. Embryonic RNAi analysis in the firebrat, Thermobia domestica: Distal-less is required to form caudal filament. J Insect Biotechnol Sericology. 2009;78(2):2_99-2_105. doi: 10.11416/jibs.78.2_99 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

