Transcriptional signature of cardiac myocyte recovery in mice and human reveals persistent upregulation of epigenetic factors
- PMID: 40472027
- PMCID: PMC12143706
- DOI: 10.1080/15592294.2025.2506625
Transcriptional signature of cardiac myocyte recovery in mice and human reveals persistent upregulation of epigenetic factors
Abstract
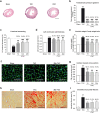
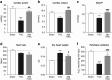
Fibrosis, cardiac remodelling, and inflammation are hallmarks of heart failure. To date, there is no available pharmacological cure for heart failure, but mechanical unloading by implantation of a left ventricular assist device (LVAD) can lead to improved cardiac function in a subset of patients. This study aimed to identify the transcriptional response of left ventricular (LV) cardiac myocytes to mechanical unloading in a mouse model of reversible LV pressure overload and in failing human hearts after LVAD implantation. We found that partial recovery of ventricular dysfunction, LV hypertrophy, and gene expression programmes occurred in mice under reversible transverse aortic constriction (rTAC). Gene expression analysis in cardiac myocytes identified a lasting repression of mitochondrial gene expression resulting in compromised fatty acid oxidation in the mouse model of reversible pressure overload and in human LV samples after LVAD therapy and a persistent upregulation of epigenetic and transcriptional regulators. These findings underpin that recovery from heart failure involves complex gene regulatory networks and that mitochondrial dysfunction remains a challenge even after mechanical unloading. Further studies are needed to investigate the functional role of these factors in reverse remodelling and recovery of failing hearts.
Keywords: Heart failure; epigenetics; mechanical unloading; mitochondria; reverse remodeling; transcriptome.
Conflict of interest statement
T.A.M. is a co-founder of Myracle Therapeutics, is on the SABs of Eikonizo Therapeutics and Revier Therapeutics, received funding from Italfarmaco for an unrelated project, and has a subcontract from Eikonizo Therapeutics for an SBIR grant from the National Institutes of Health (HL154959).
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
