Early de novo T cell expansion following SARS-CoV-2 infection predicts favourable clinical and virological outcomes
- PMID: 40472803
- PMCID: PMC12171543
- DOI: 10.1016/j.ebiom.2025.105795
Early de novo T cell expansion following SARS-CoV-2 infection predicts favourable clinical and virological outcomes
Abstract
Background: De novo T cell expansion to a novel viral infection is assumed to confer protection, but empirical evidence in humans is limited. The SARS-CoV-2 pandemic provided a unique opportunity to investigate de novo T cell-mediated protection in antigen-naïve individuals without the confounding effects of preexisting immune memory.
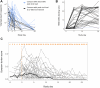
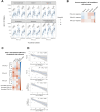
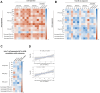
Methods: We leveraged a prospective household contact study to recruit new COVID-19 cases a median of 4 days post-SARS-CoV-2 exposure. We longitudinally enumerated SARS-CoV-2 antigen-specific functional T cell subsets using dual IFN-γ/IL-2 fluorescence-linked immunospot (FLISpot) assays. We then correlated T cell dynamics with detailed clinical and virological outcomes derived from longitudinal measurement of symptom burden and viral load.
Findings: Early expansion (day 0-7) of SARS-CoV-2-specific IFN-γ-secreting T cells correlated with lower peak viral load and symptom burden. Conversely, late T cell expansion (day 7-28) correlated with higher symptom burden. Neither pre-existing cross-reactive T cells nor early antibody induction correlated with virological outcomes.
Interpretation: These findings provide empiric evidence for early antigen-specific T cell expansion being protective against naturally acquired viral infection in humans.
Funding: This work is supported by the NIHR Health Protection Research Unit in Respiratory Infections, Imperial College London in partnership with the UK Health Security Agency (Grant number: NIHR200927; AL) and the Medical Research Council (Grant number: MR/X004058/1).
Keywords: Correlates of protection; Household contacts; SARS-CoV-2; T cell.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests. The INSTINCT study group was funded by NIHR as part of the Health Protection Research Unit in Respiratory Infections (NIHR200927).
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

