CC180 clade dynamics do not universally explain Streptococcus pneumoniae serotype 3 persistence post-vaccine: a global comparative population genomics study
- PMID: 40472804
- PMCID: PMC12172299
- DOI: 10.1016/j.ebiom.2025.105781
CC180 clade dynamics do not universally explain Streptococcus pneumoniae serotype 3 persistence post-vaccine: a global comparative population genomics study
Abstract
Background: Clonal complex 180 (CC180) is currently the major clone of serotype 3 Streptococcus pneumoniae (Spn) causing disease among children and adults worldwide. The 13-valent pneumococcal conjugate vaccine (PCV13) does not have significant efficacy against serotype 3 despite polysaccharide inclusion in the vaccine. It was hypothesized that PCV13 may effectively control Clade I of CC180 but that Clades III and IV are resistant, provoking a population shift that enables serotype 3 persistence. This has been observed in the United States, England, and Wales but not Spain. We tested this hypothesis further utilizing a dataset from Portugal to conduct our population genomics and molecular epidemiology comparative study.
Methods: We performed whole-genome sequencing (WGS) of 501 serotype 3 strains from Portugal isolated from patients with pneumococcal infections between 1999 and 2020. The draft genomes underwent phylogenetic analyses, pangenome profiling, and a genome-wide association study (GWAS). We also completed antibiotic susceptibility testing and compiled over 2600 serotype 3 multilocus sequence type 180 (MLST180) WGSs to perform global comparative genomics.
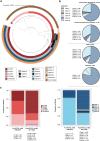
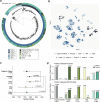
Findings: Relative to strains from all other lineages, CC180 Clades I, II, III, IV, and VI strains trend towards a decreased association with invasive disease cases compared to non-invasive pneumonia cases (binomial logistic regression, odds ratio or OR = 0.59, 95% confidence interval or CI = [0.34, 0.98], P = 0.046) and adult patients compared to paediatric patients (binomial logistic regression, OR = 0.34, 95% CI = [0.098, 0.92], P = 0.054). The serotype 3 CCs shifted post-PCV13 such that Clades I-VI comprise the majority of post-PCV13 lineages (binomial logistic regression, OR = 7.33, 95% CI = 4.36, 12.80, P < 0.0001), with Clade I representing 54% (220/404) of all post-PCV13 strains. As observed elsewhere, Clade I strains from Portugal are largely antibiotic-sensitive and carry the ΦOXC141 prophage. However, strains from Portugal and Spain, where Clade I remains dominant post-PCV13, have larger pangenomes and are associated with the presence of two genes encoding hypothetical proteins.
Interpretation: Clade I became dominant in Portugal post-PCV13, despite the burden of the prophage and antibiotic sensitivity. The additional accessory genome content may mitigate these fitness costs. Regional differences in Clade I prevalence and pangenome heterogeneity suggest that clade dynamics is not a generalizable approach to understanding serotype 3 vaccine escape.
Funding: National Institute of Child Health and Human Development, Pfizer, and Merck Sharp & Dohme.
Keywords: Clade I; Invasive pneumococcal disease; Molecular epidemiology; PCV13; Serotype 3; Streptococcus pneumonia.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests JMC is a consultant for Merck Sharp and Dohme and Pfizer in the area of pneumococcal vaccines, received research grants administered through his university from Merck and Pfizer, and received honoraria for serving on the speakers bureaus of Pfizer and Merck. MR is a consultant for Merck and Pfizer in the area of pneumococcal vaccines, received honoraria for serving on the speakers bureau of Pfizer and Merck and for serving in expert panels of Merck, and received research grants administered through his university from Merck. RM is a consultant to GlaxoSmithKline and Merck, is a named inventor and patent holder on vaccine technologies, licenced MAPS vaccine technology to GSK, received support for attending meetings from GSK, and serves on the board of directors at Corner Therapeutics and the scientific advisory boards of Amplitude Therapeutics, Corner Therapeutics, and Limmatech. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Update of
-
CC180 clade dynamics do not universally explain Streptococcus pneumoniae serotype 3 persistence post-vaccine: a global comparative population genomics study.medRxiv [Preprint]. 2024 Sep 18:2024.08.29.24312665. doi: 10.1101/2024.08.29.24312665. medRxiv. 2024. Update in: EBioMedicine. 2025 Jul;117:105781. doi: 10.1016/j.ebiom.2025.105781. PMID: 39252931 Free PMC article. Updated. Preprint.
References
-
- Troeger C., Blacker B., Khalil I.A., et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18(11):1191–1210. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

