Multi-omic spatial profiling reveals the unique SARS-CoV-2 lung microenvironment and collagen VI as a predictive biomarker in severe COVID-19
- PMID: 40473310
- PMCID: PMC12441580
- DOI: 10.1183/13993003.01699-2023
Multi-omic spatial profiling reveals the unique SARS-CoV-2 lung microenvironment and collagen VI as a predictive biomarker in severe COVID-19
Abstract
Background: While coronavirus disease 2019 (COVID-19) is primarily a respiratory infection, few studies have characterised the immune response to COVID-19 in lung tissue. We sought to understand the pathogenic role of microenvironmental interactions and the extracellular matrix in post-mortem COVID-19 lung using an integrative multi-omic approach.
Methods: Post-mortem formalin-fixed paraffin-embedded lung tissue from fatal COVID-19 and nonrespiratory death control lung underwent multi-omic evaluation by Quantseq Bulk RNA sequencing, Nanostring GeoMx spatial transcriptomics, RNAscope, multiplex immunofluorescence and immunohistochemistry, to evaluate virus distribution, immune composition and the extracellular matrix. Markers of extracellular synthesis and breakdown were measured in the serum of 215 patients with COVID-19 and 54 healthy volunteer controls using ELISA.
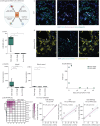
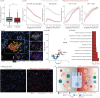
Results: We found that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection was restricted to the pneumocytes and macrophages of early-stage disease. Spatial analyses revealed an immunosuppressive virus microenvironment, enriched for PDL1+IDO1+ macrophages and depleted of T-cells. Oligoclonal T-cells in COVID-19 lung showed no enrichment of SARS-CoV-2 specific T-cell receptors. Collagen VI was upregulated and contributed to alveolar wall thickening and impaired gas exchange in COVID-19 lung. Serum from COVID-19 patients showed increased levels of PRO-C6, a marker of collagen VI synthesis, predicted mortality in hospitalised patients.
Conclusions: Our data refine the current model of respiratory COVID-19 with regard to virus distribution, immune niches and the role of the noncellular microenvironment in pathogenesis and risk stratification in COVID-19. We show that collagen deposition is an early event in the course of the disease.
Copyright ©The authors 2025.
Conflict of interest statement
Conflicts of interest: Serum ELISA assays were performed by Nordic Bioscience for no fee. J.M. Bülow Sand, M.A. Karsdal and D.J. Leeming are employees of Nordic Bioscience. Multiplex immunofluorescence (mIF) on the Akoya Phenocycler platform were performed by Akoya Biosciences for no fee. N. Nikulina, B.B. Cheikh and O. Braubach were employees of Akoya Biosciences. Initial analysis of the mIF was performed by A.T. Mayer of Enable Medicine for no fee. A.T. Mayer is president, chief scientific officer and founder of Enable Medicine. The remaining authors have no potential conflicts of interest to disclose.
Figures





Comment in
-
Multi-omics profiling in severe pneumonia: the importance of robust study design.Eur Respir J. 2025 Sep 17;66(3):2500760. doi: 10.1183/13993003.00760-2025. Print 2025 Sep. Eur Respir J. 2025. PMID: 40962430 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
