C. elegans SSNA-1 is required for the structural integrity of centrioles and bipolar spindle assembly
- PMID: 40473601
- PMCID: PMC12141670
- DOI: 10.1038/s41467-025-59939-0
C. elegans SSNA-1 is required for the structural integrity of centrioles and bipolar spindle assembly
Abstract
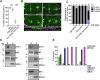
Centrioles play key roles in mitotic spindle assembly. Once assembled, centrioles exhibit long-term stability, but how stability is achieved and how it is regulated are not completely understood. In this study we show that SSNA-1, the Caenorhabditis elegans ortholog of Sjogren's Syndrome Nuclear Antigen 1, is a constituent of centrioles and centriole satellite-like structures. A deletion of ssna-1 results in the formation of extra centrioles. We show that SSNA-1 genetically interacts with the centriole stability factor SAS-1 and is required post assembly for centriole structural integrity. In SSNA-1's absence, centrioles assemble but fracture leading to extra spindle poles. However, if the efficiency of cartwheel assembly is reduced, the absence of SSNA-1 results in daughter centriole loss and monopolar spindles, indicating that the cartwheel and SSNA-1 cooperate to stabilize centrioles during assembly. Our work thus shows that SSNA-1 contributes to centriole stability during and after assembly, thereby ensuring proper centriole number.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






Update of
-
The C. elegans homolog of Sjögren's Syndrome Nuclear Antigen 1 is required for the structural integrity of the centriole and bipolar mitotic spindle assembly.bioRxiv [Preprint]. 2024 Oct 4:2024.10.03.616528. doi: 10.1101/2024.10.03.616528. bioRxiv. 2024. Update in: Nat Commun. 2025 Jun 5;16(1):5220. doi: 10.1038/s41467-025-59939-0. PMID: 39803516 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

