Muscle stem cells in Duchenne muscular dystrophy exhibit molecular impairments and altered cell fate trajectories impacting regenerative capacity
- PMID: 40473604
- PMCID: PMC12141486
- DOI: 10.1038/s41419-025-07755-1
Muscle stem cells in Duchenne muscular dystrophy exhibit molecular impairments and altered cell fate trajectories impacting regenerative capacity
Abstract
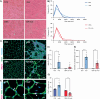
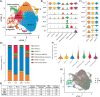
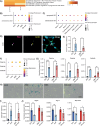
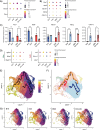
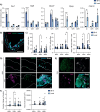
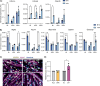
Satellite cells are muscle-resident stem cells that maintain and repair muscle. Increasing evidence supports the contributing role of satellite cells in Duchenne muscular dystrophy (DMD), a lethal degenerative muscle disease caused by loss of dystrophin. However, whether or not satellite cells exhibit dysfunction due to loss of dystrophin remains unresolved. Here, we used single-cell RNA-sequencing (scRNA-seq) to determine how dystrophin deficiency impacts the satellite cell transcriptome and cellular composition by comparing satellite cells from mdx and the more severe D2-mdx DMD mouse models. DMD satellite cells were disproportionally found within myogenic progenitor clusters and a previously uncharacterized DMD-enriched cluster. Despite exposure to different dystrophic environments, mdx and D2-mdx satellite cells exhibited overlapping dysregulation in gene expression and associated biological pathways. When comparing satellite stem cell versus myogenic progenitor populations, we identified unique dysfunctions between DMD and healthy satellite cells, including apoptotic cell death and senescence, respectively. Pseudotime analyses revealed differences in cell fate trajectories, indicating that DMD satellite cells are stalled in their differentiation capacity. In vivo regeneration assays confirmed that DMD satellite cells exhibit impaired myogenic gene expression and cell fate dynamics during regenerative myogenesis. These defects in differentiation capacity are accompanied by impaired senescence and autophagy dynamics. Finally, we demonstrate that inducing autophagy can rescue the differentiation of DMD progenitors. Our findings provide novel molecular evidence of satellite cell dysfunction in DMD, expanding on our understanding of their role in its pathology and suggesting pathways to target and enhance their regenerative capacity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: Approval for animal work was obtained from the McGill University Animal Care Committee (MCGL-8124). Housing, husbandry, and all experimental protocols for mice used in this study were performed in accordance with the guidelines established by the McGill University Animal Compliance Office, which is based on the guidelines of the Canadian Council on Animal Care.
Figures

Similar articles
-
Alterations in Notch signalling in skeletal muscles from mdx and dko dystrophic mice and patients with Duchenne muscular dystrophy.Exp Physiol. 2014 Apr;99(4):675-87. doi: 10.1113/expphysiol.2013.077255. Epub 2014 Jan 17. Exp Physiol. 2014. PMID: 24443351
-
miR-146a deficiency does not aggravate muscular dystrophy in mdx mice.Skelet Muscle. 2019 Aug 14;9(1):22. doi: 10.1186/s13395-019-0207-0. Skelet Muscle. 2019. PMID: 31412923 Free PMC article.
-
MyD88 is required for satellite cell-mediated myofiber regeneration in dystrophin-deficient mdx mice.Hum Mol Genet. 2018 Oct 1;27(19):3449-3463. doi: 10.1093/hmg/ddy258. Hum Mol Genet. 2018. PMID: 30010933 Free PMC article.
-
Satellite Cells in Muscular Dystrophy - Lost in Polarity.Trends Mol Med. 2016 Jun;22(6):479-496. doi: 10.1016/j.molmed.2016.04.002. Epub 2016 May 5. Trends Mol Med. 2016. PMID: 27161598 Free PMC article. Review.
-
Empowering Muscle Stem Cells for the Treatment of Duchenne Muscular Dystrophy.Cells Tissues Organs. 2022;211(6):641-654. doi: 10.1159/000514305. Epub 2021 Apr 28. Cells Tissues Organs. 2022. PMID: 33910206 Review.
References
-
- Relaix F, Zammit PS. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139:2845–56. - PubMed
-
- Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–56. - PubMed
MeSH terms
Substances
Grants and funding
- 175431/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- 180499/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- ECR-C4R1-2/Stem Cell Network (Le Réseau de cellules souches)
- 309857/Fonds de Recherche du Québec - Santé (Fonds de la recherche en sante du Quebec)
LinkOut - more resources
Full Text Sources
Medical

