β-propeller protein-associated neurodegeneration protein WDR45 regulates stress granule disassembly via phase separation with Caprin-1
- PMID: 40473629
- PMCID: PMC12141619
- DOI: 10.1038/s41467-025-60583-x
β-propeller protein-associated neurodegeneration protein WDR45 regulates stress granule disassembly via phase separation with Caprin-1
Abstract
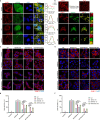
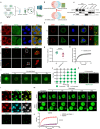
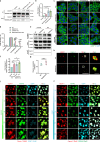
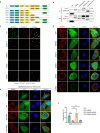
β-propeller protein-associated neurodegeneration (BPAN) is a rare X-linked neurodegenerative disorder caused by mutations in the WDR45 gene, yet its molecular mechanisms remain poorly understood. Here, we identify a role for WDR45 in stress granule (SG) disassembly, mediated through its phase separation with Caprin-1. We demonstrate that WDR45 forms gel-like condensates via its WD5 domain, which competitively displaces G3BP1 from Caprin-1 to promote SG disassembly. BPAN-associated WDR45 mutations impair condensate formation and Caprin-1 interaction, leading to delayed SG disassembly, which correlates with earlier disease onset. WDR45 depletion also exacerbates amyotrophic lateral sclerosis-associated pathological SGs, highlighting its broader relevance to neurodegenerative diseases. Using iPSC-derived midbrain neurons from a BPAN patient, we demonstrate delayed SG recovery, directly linking WDR45 dysfunction to neurodegeneration. These findings establish WDR45 as a critical regulator of SG dynamics, uncover a potential molecular basis of BPAN pathogenesis, and identify therapeutic targets for neurodegenerative diseases associated with SG dysregulation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
The Acetylation of Lysine-376 of G3BP1 Regulates RNA Binding and Stress Granule Dynamics.Mol Cell Biol. 2019 Oct 28;39(22):e00052-19. doi: 10.1128/MCB.00052-19. Print 2019 Nov 15. Mol Cell Biol. 2019. PMID: 31481451 Free PMC article.
-
Stress granule homeostasis is modulated by TRIM21-mediated ubiquitination of G3BP1 and autophagy-dependent elimination of stress granules.Autophagy. 2023 Jul;19(7):1934-1951. doi: 10.1080/15548627.2022.2164427. Epub 2023 Jan 24. Autophagy. 2023. PMID: 36692217 Free PMC article.
-
Stress granule formation helps to mitigate neurodegeneration.Nucleic Acids Res. 2024 Sep 9;52(16):9745-9759. doi: 10.1093/nar/gkae655. Nucleic Acids Res. 2024. PMID: 39106168 Free PMC article.
-
Application of stress granule core element G3BP1 in various diseases: A review.Int J Biol Macromol. 2024 Dec;282(Pt 5):137254. doi: 10.1016/j.ijbiomac.2024.137254. Epub 2024 Nov 6. Int J Biol Macromol. 2024. PMID: 39515684 Review.
-
Two Birds With One Stone: RNA Virus Strategies to Manipulate G3BP1 and Other Stress Granule Components.Wiley Interdiscip Rev RNA. 2025 Mar-Apr;16(2):e70005. doi: 10.1002/wrna.70005. Wiley Interdiscip Rev RNA. 2025. PMID: 40170442 Free PMC article. Review.
References
-
- Haack, T. B., Hogarth, P., Gregory, A., Prokisch, H. & Hayflick, S. J. BPAN: the only X-linked dominant NBIA disorder. Int Rev. Neurobiol.110, 85–90 (2013). - PubMed
-
- Ji, C., Zhao, H., Chen, D., Zhang, H. & Zhao, Y. G. β-propeller proteins WDR45 and WDR45B regulate autophagosome maturation into autolysosomes in neural cells. Curr. Biol.31, 1666–1677.e1666 (2021). - PubMed
MeSH terms
Substances
Grants and funding
- 20240484503/Beijing Nova Program
- 7244365/Natural Science Foundation of Beijing Municipality (Beijing Natural Science Foundation)
- 32471202/National Natural Science Foundation of China (National Science Foundation of China)
- 32270856/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

