Direct detection of 8-oxo-dG using nanopore sequencing
- PMID: 40473638
- PMCID: PMC12141616
- DOI: 10.1038/s41467-025-60391-3
Direct detection of 8-oxo-dG using nanopore sequencing
Abstract
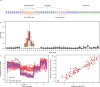
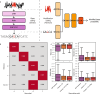
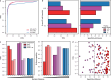
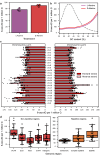
Genomic DNA is under constant oxidative damage, with 8-oxo-7,8-dihydro-2'-deoxyguanosine (8-oxo-dG) being the prominent lesion linked to mutagenesis, epigenetics, and gene regulation. Existing methods to detect 8-oxo-dG rely on indirect approaches, while nanopore sequencing enables direct detection of base modifications. A model for 8-oxo-dG detection is currently missing due to the lack of training data. Here, we develop a strategy using synthetic oligos to generate long, 8-oxo-dG context-variable DNA molecules for deep learning and nanopore sequencing. Our training approach addresses the rarity of 8-oxo-dG relative to guanine, enabling specific detection. Applied to a tissue culture model of oxidative damage, our method reveals uneven genomic 8-oxo-dG distribution, dissimilar context pattern to C>A mutations, and local 5-mC depletion. This dual measurement of 5-mC and 8-oxo-dG at single-molecule resolution uncovers new insights into their interplay. Our approach also provides a general framework for detecting other rare DNA modifications using synthetic DNA and nanopore sequencing.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.d. R. and A.M. are co-founders and directors of Cyclomics, a genomics company, they declare no competing interests. M.P-G., D.M.K.v.S., N.J.M.B., R.S., J.P.K., C.V., M.J.v.R., R.v.B., B.M.T.B., and T.B.D. declare no competing interests.
Figures
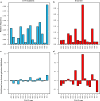
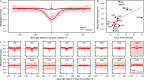
References
-
- Hush, N. S. & Cheung, A. S. Ionization potentials and donor properties of nucleic acid bases and related compounds. Chem. Phys. Lett.34, 11–13 (1975). - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

