Putative type III effector SkP48 of Bradyrhizobium sp. DOA9 encoding a SUMO protease blocks nodulation in Vigna radiata
- PMID: 40473746
- PMCID: PMC12141436
- DOI: 10.1038/s41598-025-05176-w
Putative type III effector SkP48 of Bradyrhizobium sp. DOA9 encoding a SUMO protease blocks nodulation in Vigna radiata
Abstract
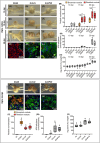
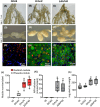
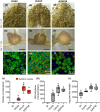
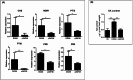
Bradyrhizobium sp. DOA9 can nodulate a wide spectrum of legumes; however, unlike other bradyrhizobia, DOA9 carries a symbiotic plasmid harboring type III secretion system (T3SS) and several effector (T3E) genes, one of which encodes a putative type III effector SkP48. Here, we demonstrated the pivotal roles of SkP48 from Bradyrhizobium sp. DOA9 in inhibiting nodulation of various Vigna species and Crotalaria juncea and suppressing nodulation efficiency of Arachis hypogaea. By contrast, the nodulation efficiency of a SkP48 mutant did not differ significantly with the DOA9 wild-type strain on Macroptilium atropurpureum and Stylosanthes hamata. The SUMO domain of SkP48 is primarily responsible for the blocking nodulation phenotype V. radiata. An evolutionary analysis revealed that the SkP48 which contains a shikimate kinase and a SUMO protease (C48 cysteine peptidase) domain, SkP48 is distinct from other effectors previously reported in other bradyrhizobia and pathogenic bacteria. Our findings suggest that the putative T3E SkP48 is a key factor suppressing nodulation and nodule organogenesis in several legumes by activation of effector-triggered immunity through salicylic acid biosynthesis induction, which is deleterious to rhizobial infection. In addition, nodulation may be modulated by the function of defensins involved in jasmonic acid signalling in V. radiata SUT1.
Keywords: Bradyrhizobium sp. DOA9; Vigna radiata; SkP48; Symbiosis; T3SS.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
-
- Janczarek, M., Rachwał, K., Marzec, A., Grzadziel, J. & Palusińska-Szysz, M. Signal molecules and cell-surface components involved in early stages of the legume-rhizobium interactions. Appl. Soil. Ecol.85, 94–113 (2015). - DOI
MeSH terms
Substances
Grants and funding
- TRG6280006/National Research Council of Thailand (NRCT) and Suranaree University of Technology / The Thailand Research Fund project
- B13F660067/NSRF via the Program Management Unit for Human Resources & Institutional Development, Research, and Innovation (PMU-B)
- B13F660067/NSRF via the Program Management Unit for Human Resources & Institutional Development, Research, and Innovation (PMU-B)
- B13F660067/NSRF via the Program Management Unit for Human Resources & Institutional Development, Research, and Innovation (PMU-B)
- B13F660067/NSRF via the Program Management Unit for Human Resources & Institutional Development, Research, and Innovation (PMU-B)
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- 195582/National Science, Research, and Innovation Fund (NSRF)
- 195582/National Science, Research, and Innovation Fund (NSRF)
- 195582/National Science, Research, and Innovation Fund (NSRF)
- 195582/National Science, Research, and Innovation Fund (NSRF)
- ("ET-Nod"; ANR-20-CE20-0012)/Grant from the French National Research Agency
LinkOut - more resources
Full Text Sources

