Combined dendritic cell and anti-TIGIT immunotherapy potentiates adaptive NK cells against HIV-1
- PMID: 40473838
- PMCID: PMC12254423
- DOI: 10.1038/s44321-025-00255-x
Combined dendritic cell and anti-TIGIT immunotherapy potentiates adaptive NK cells against HIV-1
Abstract
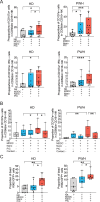
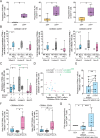
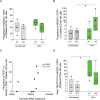
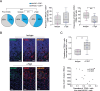
Natural Killer (NK) cells are promising candidates for targeting persistently infected CD4 + T cells in people with HIV-1 (PWH). However, chronicity of HIV-1 infection impairs NK cell functionality, requiring additional strategies to potentiate their cytotoxic activity. This study demonstrates that dendritic cells primed with nanoparticles containing Poly I:C (Nano-PIC-MDDC) enhance the natural cytotoxic function of NK cells from effective responder PWH. These NK cells exhibit increased proportions of NKG2C+ cell subsets capable of eliminating HIV-1 infected CD4 + T cells through the TRAIL receptor. In contrast, in non-responder PWH, elevated expression of the inhibitory receptor TIGIT is associated with reduced frequencies of NKG2C + NK cells and diminished TRAIL expression. TIGIT blockade restores cytotoxicity of NK cells from non-responder PWH against HIV-1-infected cells by upregulating TRAIL. Furthermore, combining Nano-PIC-MDDC-primed NK cells with anti-TIGIT immunotherapy in humanized NSG mice reduces the expansion of HIV-1 infected cells, preserves NKG2C + NK cell precursors and increases TRAIL expression in tissue. Collectively, these findings support the combined use of Nano-PIC-MDDC and TIGIT blockade as a promising immunotherapeutic strategy toward an HIV-1 cure.
Keywords: Dendritic Cell; HIV; Natural Killer; TIGIT; TRAIL.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
- PID2023-149541OB-I00/'la Caixa' Foundation ('la Caixa')
- PI22/01120/Centro de Investigación Biotecnológica en Red de Enfermedades Infecciosas (CIBERINFEC)
- FPI UAM/Universidad Autónoma de Madrid (UAM)
- P2022/BMD7209/Comunidad de Madrid (Community of Madrid)
- PRE2022-104516/MEC | Agencia Estatal de Investigación (AEI)
- 2017-T1/BMD-5396/Comunidad de Madrid (Community of Madrid)
- RYC2018-024374-I/Ministerio de Asuntos Económicos y Transformación Digital, Gobierno de España (MINECO)
- HR20-00218/'la Caixa' Foundation ('la Caixa')
- PID2022-138880OB-I00; PDC2021-121238-I00/Ministerio de Ciencia e Innovación (MCIN)
- CB21/13/00107/Centro de Investigación Biotecnológica en Red de Enfermedades Infecciosas (CIBERINFEC)
- GLD24/00117/Gilead Foundation
- RTI2018-097485-A-I00; PID2021-127899OB-I00; CNS2023-144841/Ministerio de Ciencia e Innovación (MCIN)
- L60 MD007209/MD/NIMHD NIH HHS/United States
- LCF/PR/HR23/52430018/'la Caixa' Foundation ('la Caixa')
LinkOut - more resources
Full Text Sources
Medical
Research Materials

