Molecular dynamics guided identification of a brighter variant of superfolder Green Fluorescent Protein with increased photobleaching resistance
- PMID: 40473962
- PMCID: PMC12141695
- DOI: 10.1038/s42004-025-01573-4
Molecular dynamics guided identification of a brighter variant of superfolder Green Fluorescent Protein with increased photobleaching resistance
Abstract
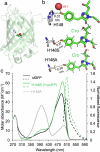
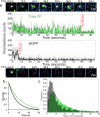
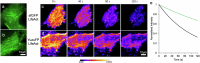
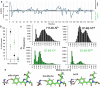
Fluorescent proteins (FPs) are a crucial tool for cell imaging, but with developments in fluorescence microscopy and researcher requirements there is still a need to develop brighter versions that remain fluorescent for longer. Using short time-scale molecular dynamics-based modelling to predict changes in local chromophore interaction networks and solvation, we constructed an Aequorea victoria GFP (avGFP) variant called YuzuFP that is 1.5 times brighter than the starting superfolding variant (sfGFP) with a near 3-fold increased resistance to photobleaching in situ. YuzuFP contained a single mutation that replaces the chromophore interacting residue H148 with a serine. Longer time scale molecular dynamics revealed the likely mechanism of action: S148 forms more persistent H-bond with the chromophore phenolate group and increases the residency time of an important water molecule. As demonstrated by live cell imaging, YuzuFP not only offers a timely upgrade as a useful green-yellow avGFP for cell imaging applications over longer time scales, but it also provides a basic scaffold for future avGFP engineering efforts.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interesting.
Figures
Similar articles
-
Amino acid residue at the 165th position tunes EYFP chromophore maturation. A structure-based design.Comput Struct Biotechnol J. 2021 May 11;19:2950-2959. doi: 10.1016/j.csbj.2021.05.017. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34136094 Free PMC article.
-
Hue-shifted monomeric variants of Clavularia cyan fluorescent protein: identification of the molecular determinants of color and applications in fluorescence imaging.BMC Biol. 2008 Mar 6;6:13. doi: 10.1186/1741-7007-6-13. BMC Biol. 2008. PMID: 18325109 Free PMC article.
-
Cyan fluorescent proteins derived from mNeonGreen.Protein Eng Des Sel. 2022 Feb 17;35:gzac004. doi: 10.1093/protein/gzac004. Protein Eng Des Sel. 2022. PMID: 35417013 Free PMC article.
-
Advances in fluorescent protein technology.J Cell Sci. 2007 Dec 15;120(Pt 24):4247-60. doi: 10.1242/jcs.005801. J Cell Sci. 2007. PMID: 18057027 Review.
-
Cell-based assays in practice: cell markers from autofluorescent proteins of the GFP-family.Comb Chem High Throughput Screen. 2008 Sep;11(8):602-9. doi: 10.2174/138620708785739880. Comb Chem High Throughput Screen. 2008. PMID: 18795879 Review.
References
-
- Shaner, N. C., Steinbach, P. A. & Tsien, R. Y. A guide to choosing fluorescent proteins. Nat. Methods2, 905–909 (2005). - PubMed
-
- Zimmer, M. Green fluorescent protein (GFP): applications, structure, and related photophysical behavior. Chem. Rev.102, 759–781 (2002). - PubMed
-
- Duwé, S. & Dedecker, P. Optimizing the fluorescent protein toolbox and its use. Curr. Opin. Biotechnol.58, 183–191 (2019). - PubMed
Grants and funding
- BB/Z514913/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/Y008537/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- EP/V048147/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
- 824060/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

