High-definition spatial transcriptomic profiling of immune cell populations in colorectal cancer
- PMID: 40473992
- PMCID: PMC12165841
- DOI: 10.1038/s41588-025-02193-3
High-definition spatial transcriptomic profiling of immune cell populations in colorectal cancer
Abstract
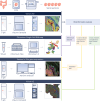
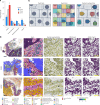
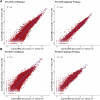
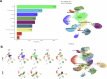
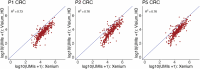
A comprehensive understanding of cellular behavior and response to the tumor microenvironment (TME) in colorectal cancer (CRC) remains elusive. Here, we introduce the high-definition Visium spatial transcriptomic technology (Visium HD) and investigate formalin-fixed paraffin-embedded human CRC samples (n = 5). We demonstrate the high sensitivity, single-cell-scale resolution and spatial accuracy of Visium HD, generating a highly refined whole-transcriptome spatial profile of CRC samples. We identify transcriptomically distinct macrophage subpopulations in different spatial niches with potential pro-tumor and anti-tumor functions via interactions with tumor and T cells. In situ gene expression analysis validates our findings and localizes a clonally expanded T cell population close to macrophages with anti-tumor features. Our study demonstrates the power of high-resolution spatial technologies to understand cellular interactions in the TME and paves the way for larger studies that will unravel mechanisms and biomarkers of CRC biology, improving diagnosis and disease management strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: All authors are current or former employees or shareholders of 10x Genomics.
Figures








References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

