A capsular myofibroblastic niche maintains hematopoietic stem cells in the spleen
- PMID: 40474007
- PMCID: PMC12264044
- DOI: 10.1038/s44318-025-00477-2
A capsular myofibroblastic niche maintains hematopoietic stem cells in the spleen
Abstract
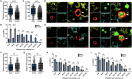
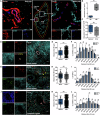
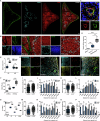
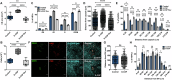
The spleen is a key site for extramedullary hematopoiesis that hosts a rare population of functional hematopoietic stem cells (HSCs). While the microenvironment that supports extramedullary hematopoiesis response has gained interest, a niche for splenic HSCs at steady-state remains undescribed. Here, we have uncovered a red-pulp-specific, myofibroblastic niche that supports murine splenic HSCs within a ≈ 200-μm-wide capsular zone. Detailed spatial-distribution and perturbation analysis showed the importance of myofibroblasts in maintaining HSCs in a quiescent state. Unlike reported for the adult bone marrow, the HSCs in splenic niche were not spatially associated with vascular components. G-CSF-mediated chemokine alteration and 5-FU-induced proliferation resulted in HSCs shifts away from the splenic capsule. Interestingly, upon regaining quiescence, the HSCs re-occupied niches close to capsular myofibroblasts. Proteomic interactome profiles confirmed the relevance of capsular myofibroblasts for splenic HSCs and identified potential niche regulators of HSC maintenance. Together, this study demonstrates a dynamic HSC localization in the spleen and its niche context at homeostasis and under stress. It offers a model to uncover novel regulators crucial for HSC function.
Keywords: Hematopoietic Niche; Hematopoietic Stem Cells; Myofibroblasts; Proteomics; Spleen.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures









Similar articles
-
Exploring Epigenetic Complexity in Regulation of Hematopoietic Stem Cells Niche: A Mechanistic Journey from Normal to Malignant Hematopoiesis.Adv Exp Med Biol. 2025;1483:49-67. doi: 10.1007/5584_2024_846. Adv Exp Med Biol. 2025. PMID: 39841383 Review.
-
Bone Marrow Niche in Cardiometabolic Disease: Mechanisms and Therapeutic Potential.Circ Res. 2025 Jan 31;136(3):325-353. doi: 10.1161/CIRCRESAHA.124.323778. Epub 2025 Jan 30. Circ Res. 2025. PMID: 39883790 Review.
-
CK2α Deletion in the Hematopoietic Compartment Shows a Mild Alteration in Terminally Differentiated Cells and the Expansion of Stem Cells.Cells. 2025 Jun 24;14(13):963. doi: 10.3390/cells14130963. Cells. 2025. PMID: 40643484 Free PMC article.
-
KLF4 enhances transplantation-induced hematopoiesis by inhibiting TLRs and noncanonical NFκB signaling at a steady state.Exp Hematol. 2025 Apr;144:104730. doi: 10.1016/j.exphem.2025.104730. Epub 2025 Feb 1. Exp Hematol. 2025. PMID: 39900173
-
Megakaryocytic IGF1 coordinates activation and ferroptosis to safeguard hematopoietic stem cell regeneration after radiation injury.Cell Commun Signal. 2024 May 27;22(1):292. doi: 10.1186/s12964-024-01651-5. Cell Commun Signal. 2024. PMID: 38802843 Free PMC article.
References
-
- Bennett M, Pinkerton PH, Cudkowicz G, Bannerman RM (1968) Hemopoietic progenitor cells in marrow and spleen of mice with hereditary iron deficiency anemia. Blood 32:908–921 - PubMed
MeSH terms
Grants and funding
- Intramural support/Indian Institute of Science Education and Research Thiruvananthapuram (IISER TVM)
- CRG/2022/000834/DST | Science and Engineering Research Board (SERB)
- SR/FST/LS-II/2018/217/Department of Science and Technology, Ministry of Science and Technology, India (DST)
- G00003688/Startup grant, UAE University
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

