Genome-wide characterization and their roles in abiotic stress responses of HD-Zip transcription factors in pepper
- PMID: 40474057
- PMCID: PMC12139069
- DOI: 10.1186/s12870-025-06798-y
Genome-wide characterization and their roles in abiotic stress responses of HD-Zip transcription factors in pepper
Abstract
Background: Capsicum annuum is a globally cultivated crop of significant agricultural and economic importance. However, its productivity and fruit quality are frequently challenged by a range of abiotic stresses. The HD-Zip (Homeodomain-Leucine Zipper) gene family, unique to plants, is known to play pivotal regulatory roles in abiotic stress adaptation, yet its functional roles in pepper remain largely unexplored.
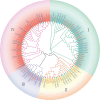
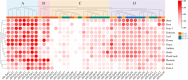
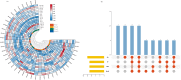
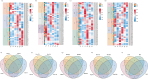
Results: This study systematically analyzed the HD-Zip gene family in pepper through bioinformatics, expression profiling, and responses to abiotic stresses and phytohormones to elucidate their roles in stress tolerance. Results revealed 40 HD-Zip transcription factors unevenly distributed across 12 chromosomes, encoding proteins ranging from 211 to 842 amino acids. Subcellular localization predictions indicated nuclear localization for all members, with a subset also showing cytoplasmic localization. Collinearity analysis demonstrated that CaHD-Zip gene expansion was predominantly driven by segmental duplication, with high conservation across dicotyledons. Promoter regions of CaHD-Zip genes were enriched in cis-regulatory elements associated with light and hormonal responses, as well as stress adaptation. Tissue-specific and developmental stage-dependent expression patterns highlighted functional diversification within the family. Notably, some members were specifically induced by abiotic stresses (cold, heat, drought, and salt) and stress-related phytohormones (ABA, MeJA, ET, and SA), suggesting their involvement in stress signaling. Strikingly, CaHD-Zip18 and CaHD-Zip29 were significantly upregulated under all four stresses, implicating them as core regulators of multi-stress responses. Subsequent stress simulation assays and qRT-PCR validation confirmed the reliability of transcriptomic findings.
Conclusion: This study delivers the first systematic exploration of HD-Zip transcription factors in Capsicum annuum under abiotic stress, providing foundational knowledge and candidate genes for improving stress resilience in pepper breeding programs.
Keywords: Capsicum annuum; HD-Zip gene family; Abiotic stress; Gene expression.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Characterization and Expression Analysis of β-Glucosidase Gene Under Abiotic Stresses in Pepper (Capsicum annuum L.).Genes (Basel). 2025 Jul 27;16(8):889. doi: 10.3390/genes16080889. Genes (Basel). 2025. PMID: 40869937 Free PMC article.
-
Knockdown of CaHSP60-6 confers enhanced sensitivity to heat stress in pepper (Capsicum annuum L.).Planta. 2019 Dec;250(6):2127-2145. doi: 10.1007/s00425-019-03290-4. Epub 2019 Oct 12. Planta. 2019. PMID: 31606756
-
Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.).BMC Plant Biol. 2015 Jun 19;15:151. doi: 10.1186/s12870-015-0512-7. BMC Plant Biol. 2015. PMID: 26088319 Free PMC article.
-
The role of HD-Zip class I transcription factors in plant response to abiotic stresses.Physiol Plant. 2019 Dec;167(4):516-525. doi: 10.1111/ppl.12965. Epub 2019 Apr 17. Physiol Plant. 2019. PMID: 30851063 Review.
-
HD-ZIP Gene Family: Potential Roles in Improving Plant Growth and Regulating Stress-Responsive Mechanisms in Plants.Genes (Basel). 2021 Aug 17;12(8):1256. doi: 10.3390/genes12081256. Genes (Basel). 2021. PMID: 34440430 Free PMC article. Review.
References
-
- Zhou C, Zhang W, Yu B, Yang H, Zhao Q, Wang Y, Sun K, Lakshmanan P, Chen X, Zou C. Global analysis Of spatio-temporal variation in mineral nutritional quality Of pepper (Capsicum spp.) Fruit and its regulatory Variables: A Meta-Analysis. Food Res Int. 2024;193:114855. 10.1016/j.foodres.2024.114855. - PubMed
-
- Tripodi P, Kumar S. The Capsicum Crop: An introduction. in the Capsicum Genome; Ramchiary, N., Kole, C., Eds.; Springer International Publishing: Cham, 2019; pp 1–8. 10.1007/978-3-319-97217-6_1.
-
- Moseley A, Laipply K, Rubinstein J. Capsaicin Mediates Remote Ischemic Pre-Conditioning to Explain Improved Cardiovascular Mortality With Chili Pepper Intake. J Am Coll Cardiol. 2020;75:1865. 10.1016/j.jacc.2020.01.060. - PubMed
-
- Parvez G M M. Current advances in pharmacological activity and toxic effetcs of various capsicum species. Int J Pharm Sci Res. 8. 10.13040/IJPSR.0975-8232.8(5).1900-12.
-
- Baenas N, Belović M, Ilic N, Moreno DA, García-Viguera C. Industrial Use of Pepper (Capsicum Annum L.) derived products: technological benefits and biological advantages. Food Chem. 2019;274:872–85. 10.1016/j.foodchem.2018.09.047. - PubMed
MeSH terms
Substances
Grants and funding
- 2023YFD2300704/The National Key Research and Development Program of China
- 2023YFD2300704/The National Key Research and Development Program of China
- 2023YFD2300704/The National Key Research and Development Program of China
- 2023YFD2300704/The National Key Research and Development Program of China
- CARS-23-B05/CARS
LinkOut - more resources
Full Text Sources

