Tumor suppressor SLC9A2 inhibits colorectal cancer metastasis and reverses immunotherapy resistance by suppressing angiogenesis
- PMID: 40474298
- PMCID: PMC12139321
- DOI: 10.1186/s13046-025-03422-7
Tumor suppressor SLC9A2 inhibits colorectal cancer metastasis and reverses immunotherapy resistance by suppressing angiogenesis
Abstract
Background: Colorectal cancer (CRC) is a common and aggressive malignancy that frequently metastasizes to the liver, presenting significant therapeutic challenges. Despite its clinical importance, the mechanisms underlying CRC liver metastasis and resistance to immune therapy remain poorly understood. In this study, we aimed to investigate the molecular mechanisms driving CRC metastasis using a novel approach, which includes the establishment of highly metastatic CRC cell lines.
Methods: To explore the role of specific genes in CRC liver metastasis, we developed two highly metastatic CRC cell lines (LoVo-Hm and HCT116-Hm) by applying sustained selective pressure to primary CRC cells. RNA sequencing was performed to identify differentially expressed genes in these metastatic cells. Additionally, we conducted assays for cell migration, invasion, angiogenesis, and ELISA to evaluate VEGFA production, all to confirm the functional role of SLC9A2. Our findings were further validated in human CRC tissue samples and publicly available datasets to assess the clinical relevance of the identified targets.
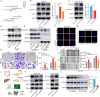
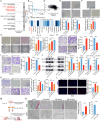
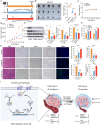
Results: Our analysis revealed a significant downregulation of SLC9A2 in the highly metastatic CRC cell lines. Mechanistically, we found that SLC9A2 inhibits epithelial-mesenchymal transition (EMT) and metastasis by suppressing the STAT3 signaling pathway. Moreover, SLC9A2 reduces VEGFA secretion, normalizing tumor vasculature and reshaping the tumor microenvironment (TME), which ultimately enhances anti-tumor immunity. Comparative analysis of CRC tissue samples showed reduced SLC9A2 expression in tumor tissues compared to adjacent normal tissues, with a negative correlation to TNM staging. Importantly, higher SLC9A2 expression was associated with better treatment responses in immunotherapy cohorts.
Conclusion: These findings highlight the critical role of SLC9A2 in regulating metastasis, angiogenesis, and TME remodeling in CRC. By modulating the STAT3 pathway and tumor vasculature, SLC9A2 emerges as a potential prognostic biomarker and therapeutic target. Targeting SLC9A2 may enhance immune responses and improve treatment outcomes in CRC, offering a promising avenue for future therapeutic strategies.
Keywords: Angiogenesis; Colorectal Cancer; Liver metastasis, immunotherapy; SLC9A2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Animal experiment is in accordance with the regulations of the Animal Care and Use Committee of the Peking University Cancer Hospital (EAEC-2023-13). Consent for publication: All authors agree to be published. Competing interests: The authors declare no competing interests.
Figures





References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–63. - PubMed
-
- Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73(3):233–54. - PubMed
-
- Tsilimigras DI, Brodt P, Clavien PA, Muschel RJ, D’Angelica MI, Endo I, Parks RW, Doyle M, de Santibañes E, Pawlik TM. Liver metastases. Nat Rev Dis Primers. 2021;7(1):27. - PubMed
MeSH terms
Substances
Grants and funding
- No. 2024T170034/the China Postdoctoral Science Foundation
- No. BJCH2024GG03/the Science Foundation of Peking University Cancer Hospital
- No. 82403443/National Natural Science Foundation of China
- No. 82373252/National Natural Science Foundation of China
- No. 2023SGGZ071/Scientific Research Fund for the Demonstration Project of Public Hospital Reform and Quality Development (Gastrointestinal Tumour) that is approved by Peking University Cancer Hospital (Inner Mongolia Campus)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

