Measuring natural selection on the transcriptome
- PMID: 40474340
- PMCID: PMC12329170
- DOI: 10.1111/nph.70287
Measuring natural selection on the transcriptome
Abstract
The level and pattern of gene expression is increasingly recognized as a principal determinant of plant phenotypes and thus of fitness. The estimation of natural selection on the transcriptome is an emerging research discipline. We here review recent progress and consider the challenges posed by the high dimensionality of the transcriptome for the multiple regression methods routinely used to characterize selection in field experiments. We consider several different methods, including classical multivariate statistical approaches, regularized regression, latent factor models, and machine learning, that address the fact that the number of traits potentially affecting fitness (each expressed gene) can greatly exceed the number of plants that researchers can reasonably monitor in a field study. While such studies are currently few, extant data are sufficient to illustrate several of these approaches. With additional methodological development coupled with applications to a broader range of species, we believe prospects are favorable for directly characterizing selection on gene expression within natural plant populations.
Keywords: RNA‐seq; coexpression networks; eQTL; fitness; natural selection; transcriptomes.
© 2025 The Author(s). New Phytologist © 2025 New Phytologist Foundation.
Conflict of interest statement
None declared.
Figures
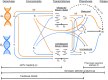
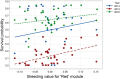
References
-
- Arnold SJ. 1983. Morphology, performance and fitness. American Zoologist 23: 347–361.
-
- Blows MW, Allen SL, Collet JM, Chenoweth SF, McGuigan K. 2015. The phenome‐wide distribution of genetic variance. The American Naturalist 186: 15–30. - PubMed
-
- Campbell‐Staton SC, Cheviron ZA, Rochette N, Catchen J, Losos JB, Edwards SV. 2017. Winter storms drive rapid phenotypic, regulatory, and genomic shifts in the green anole lizard. Science 357: 495–498. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

