Predicting Resistance and Survival of HCC Patients Post-HAIC: Based on Shapley Additive exPlanations and Machine Learning
- PMID: 40475092
- PMCID: PMC12140075
- DOI: 10.2147/JHC.S523806
Predicting Resistance and Survival of HCC Patients Post-HAIC: Based on Shapley Additive exPlanations and Machine Learning
Abstract
Purpose: To establish prediction models using Shapley Additive exPlanations (SHAP) and multiple machine learning (ML) algorithms to identify clinical features influencing hepatic arterial infusion chemotherapy (HAIC) resistance and survival in patients with hepatocellular carcinoma (HCC).
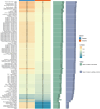
Patients and methods: We recruited 286 patients with unresectable HCC who underwent HAIC. Patients were divided into training and validation datasets (7:3 ratio). eXtreme Gradient Boosting (XGBoost) was used to build the preliminary resistance prediction model. The SHAP values explained the importance of the clinical features. Recursive Feature Elimination with Cross-Validation (RFECV) was used to select the optimum number of features. Seven ML methods were used to construct further resistance prediction models, and ten ML algorithms were employed to establish the survival prognosis models.
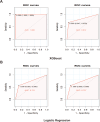
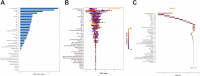
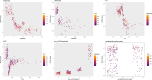
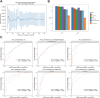
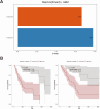
Results: The areas under the curve (AUC) of the XGBoost model were 1.000 and 0.812 for the training and validation groups, respectively. SHAP identified 27 of the 38 clinical features affecting resistance, with pre-HAIC treatment being the main factor. RFECV showed the best model performance with six features (pre-HAIC treatment, tumor size, HBV DNA, alkaline phosphatase (AKP), prothrombin time (PT), and portal vein tumor thrombosis (PVTT)). Random Forest had the best performance among the seven ML algorithms (AUC=0.935 for training, AUC=0.876 for validation). The combination of Stepcox [forward] and Gradient Boosting Machine was the best for predicting survival (AUC=0.98 in training, AUC=0.83 in validation). Based on the above clinical characteristics, patients were categorized into high-risk and low-risk groups based on the median risk score, and it was found that these characteristics also performed well in the prognostic model for predicting the survival of patients with HCC.
Conclusion: Pre-HAIC treatment, tumor size, HBV DNA, AKP, PT, and PVTT are effective predictors of post-HAIC resistance and survival in patients with unresectable advanced HCC.
Keywords: hepatocellular carcinoma outcomes; interpretable AI; prognostic modeling; treatment response prediction.
© 2025 Yao et al.
Conflict of interest statement
The authors report no conflicts of interest in this work.
Figures


Similar articles
-
Prognosis prediction and risk stratification of transarterial chemoembolization or intraarterial chemotherapy for unresectable hepatocellular carcinoma based on machine learning.Eur Radiol. 2024 Aug;34(8):5094-5107. doi: 10.1007/s00330-024-10581-2. Epub 2024 Jan 30. Eur Radiol. 2024. PMID: 38291256 Free PMC article.
-
Automatic prediction of hepatic arterial infusion chemotherapy response in advanced hepatocellular carcinoma with deep learning radiomic nomogram.Eur Radiol. 2023 Dec;33(12):9038-9051. doi: 10.1007/s00330-023-09953-x. Epub 2023 Jul 27. Eur Radiol. 2023. PMID: 37498380
-
Interpretable machine learning model for predicting post-hepatectomy liver failure in hepatocellular carcinoma.Sci Rep. 2025 May 3;15(1):15469. doi: 10.1038/s41598-025-97878-4. Sci Rep. 2025. PMID: 40316613 Free PMC article.
-
Comparative efficacy and safety of multimodality treatment for advanced hepatocellular carcinoma with portal vein tumor thrombus: patient-level network meta-analysis.Front Oncol. 2024 Feb 16;14:1344798. doi: 10.3389/fonc.2024.1344798. eCollection 2024. Front Oncol. 2024. PMID: 38434681 Free PMC article.
-
Systematic review of hepatic arterial infusion chemotherapy versus sorafenib in patients with hepatocellular carcinoma with portal vein tumor thrombosis.J Gastroenterol Hepatol. 2020 Aug;35(8):1277-1287. doi: 10.1111/jgh.15010. Epub 2020 Mar 3. J Gastroenterol Hepatol. 2020. PMID: 32052876
References
LinkOut - more resources
Full Text Sources

