This is a preprint.
A multi-omics resource of B cell activation reveals genetic mechanisms for immune-mediated diseases
- PMID: 40475139
- PMCID: PMC12140513
- DOI: 10.1101/2025.05.22.25328104
A multi-omics resource of B cell activation reveals genetic mechanisms for immune-mediated diseases
Abstract
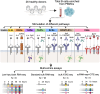
Most genetic variants that confer risk of complex immune-mediated diseases (IMDs) affect gene regulation in specific cell types. Their target genes and focus cell types are often unknown, partially because some effects are hidden in untested cell states. B cells play central roles in IMDs, including autoimmune, allergic, infectious, and cancer-related diseases. Despite this established importance, B cell activation states are underrepresented in functional genomics studies. In this study, we obtained B cells from 26 healthy female donors and stimulated them in vitro with six activation conditions targeting key pathways: the B cell receptor (BCR), Toll-like receptor 7 (TLR7), TLR9, CD40, and a cocktail that promotes differentiation into double negative 2 (DN2) IgD- CD27- CD11c+ CD21- B cells, a likely pathogenic subset implicated in autoimmunity and infection. We profiled up to 24 B cell activation states and up to 5 control conditions using RNA-seq, single-cell RNA-seq with surface protein markers (CITE-seq), and ATAC-seq. We characterize how IMD-associated genes respond to stimuli and group into distinct functional programs. High-depth RNA-seq data reveals widespread splicing effects during B cell activation. Using single-cell data, we describe stimulus-dependent B cell fates. Chromatin data reveal transcription factors likely involved in B cell activation, and activation-dependent open chromatin regions that are enriched in IMD genetic risk. We experimentally validate a lupus risk variant in a stimulus-specific open chromatin region that regulates TNFSF4 expression, highlighting the relevance of studying B cell activation to elucidate disease association. These data are shared via an interactive browser that can be used to query the dynamics of gene regulation and B cell differentiation during activation by different stimuli, enhancing further investigation of B cells and their role in IMDs: https://mgalab.shinyapps.io/bcellactivation.
Figures





References
-
- Jenks S.A., Cashman K.S., Zumaquero E., Marigorta U.M., Patel A.V., Wang X., Tomar D., Woodruff M.C., Simon Z., Bugrovsky R., et al. (2018). Distinct Effector B Cells Induced by Unregulated Toll-like Receptor 7 Contribute to Pathogenic Responses in Systemic Lupus Erythematosus. Immunity 49, 725–739. 10.1016/j.immuni.2018.08.015. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
