This is a preprint.
Aperiodic neural dynamics define a novel signature of glioma-induced excitation-inhibition dysregulation
- PMID: 40475422
- PMCID: PMC12139770
- DOI: 10.1101/2025.05.23.655626
Aperiodic neural dynamics define a novel signature of glioma-induced excitation-inhibition dysregulation
Abstract
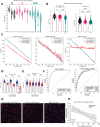
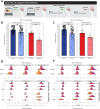
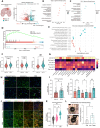
Diffuse gliomas remodel neuronal circuits with prognostic and therapeutic significance for patients. Electrophysiologic measures of cortical excitability hold promise for monitoring disease progression and evaluating therapeutic responses. The power law exponent (aperiodic slope) reflects the balance between excitatory and inhibitory activity within neuronal networks, a critical aspect of normal brain function often disrupted in neurological conditions. Despite its potential, the significance of the aperiodic slope in glioma-infiltrated tissue and its underlying cellular processes has not been fully investigated. Here, we integrate multimodal electrophysiological analysis with transcriptomic profiling to analyze the aperiodic slope in both normal and glioma-infiltrated cortex. We determine that glioma infiltration induces a flattening of the aperiodic slope, indicating a shift toward excitation dominance that varies according to tumor subtype and correlates with impairments in semantic naming. Single-nucleus RNA sequencing revealed that cortical regions with flat aperiodic slope exhibit transcriptional programs enriched in glutamatergic signaling, membrane depolarization, and excitatory synaptic transmission. The aperiodic slope responds to pharmacologically induced changes in cortical inhibition during propofol administration, a GABAA agonist. Our results establish the aperiodic slope as a robust biomarker of glioma-associated excitation-inhibition imbalance, with potential applications in tumor classification and treatment monitoring.
Keywords: Aperiodic slope; excitation-inhibition balance; glioblastoma; glioma; isocitrate dehydrogenase mutant glioma; snRNA-seq; transcriptomics.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures


References
-
- Epstein F. H., Lipton S. A. & Rosenberg P. A. Excitatory Amino Acids as a Final Common Pathway for Neurologic Disorders. New England Journal of Medicine 330, 613–622 (1994). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
