This is a preprint.
Adaptive Evolution of Gene Regulatory Networks in Mammalian Neocortical Neurons
- PMID: 40475427
- PMCID: PMC12139996
- DOI: 10.1101/2025.05.20.652233
Adaptive Evolution of Gene Regulatory Networks in Mammalian Neocortical Neurons
Abstract
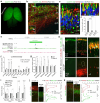
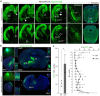
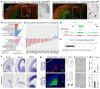
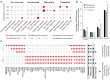
Mammals have evolved a plethora of adaptations that have enabled them to thrive in diverse environments. Among the most significant is the emergence of a more complex brain, exemplified by the dramatic transformation of the dorsal cortex from a single layer of excitatory projection neurons (ExNs) in ancestors to a multilayered cerebral neocortex enriched with diverse intratelencephalic (IT) and extratelencephalic (ET) ExN subtypes. These ExNs established specialized projection systems, such as the corticospinal tract and corpus callosum, enhancing brain connectivity and functionality. However, the evolutionary mechanisms underlying these mammalian-specific adaptations remain elusive. By comparing the landscape of gene expression and cis-regulatory elements (CREs) in mouse ExN subtypes and by cross-species examination of mammalian and non-mammalian CREs, we identified mammalian-specific CREs and expression patterns. The mammalian-specific CREs include a subset bound by ZBTB18 that are associated with genes defining IT and ET subtypes and connectivity. Both ZBTB18 and these target genes have previously been implicated in intellectual disability and autism. Deletion of Zbtb18 in mouse ExNs dysregulated target gene expression, reduced molecular diversity, diminished corticospinal and callosal projections, and increased intrahemispheric cortico-cortical association projections to the prefrontal cortex, resembling features of non-mammalian dorsal pallium. Interestingly, ZBTB18 binding motifs are highly enriched in callosally projecting IT-biased CREs, where they show higher conservation specifically in mammals. This study uncovers critical components and mammalian-specific evolutionary adaptations within a regulatory node essential for neocortical ExN identity and connectivity, with implications for neurodevelopmental and neuropsychiatric disorders.
Figures

References
-
- Striedter G. & Northcutt R. Brains Through Time: A Natural History of VertebratesA Natural History of Vertebrates.(2019).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
