This is a preprint.
Inflamed Microglia like Macrophages in the Central Nervous System of Prodromal Parkinson's Disease
- PMID: 40475455
- PMCID: PMC12139992
- DOI: 10.1101/2025.05.16.654530
Inflamed Microglia like Macrophages in the Central Nervous System of Prodromal Parkinson's Disease
Abstract
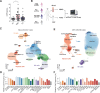
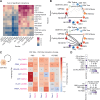
We investigated the role of inflammation in the pathogenesis of prodromal Parkinson's Disease (PD), performing single-cell RNAseq analysis of cerebrospinal fluid (CSF) and blood from 111 individuals, comparing control subjects with early prodromal PD and later PD to patients with multiple sclerosis (MS). Surprisingly, we identified a pleocytosis in the CSF, most pronounced in patients with early PD. Single-cell RNAseq revealed increases in CSF-specific microglia-like macrophages expressing JAK-STAT and TNFα signaling signatures in prodromal PD, with a lack of T cell activation in the CSF. The CSF macrophages exhibited similar transcriptional profiles to dural macrophages from human α-synuclein-expressing PD model mice. These findings uncover a myeloid-mediated TNFα inflammatory process in the CNS of patients with prodromal PD, suggesting a novel pathological mechanism in disease etiology.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
