This is a preprint.
ChromMovie: A Molecular Dynamics Approach for Simultaneous Modeling of Chromatin Conformation Changes from Multiple Single-Cell Hi-C Maps
- PMID: 40475498
- PMCID: PMC12139908
- DOI: 10.1101/2025.05.16.654550
ChromMovie: A Molecular Dynamics Approach for Simultaneous Modeling of Chromatin Conformation Changes from Multiple Single-Cell Hi-C Maps
Abstract
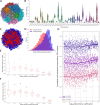
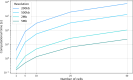
The development of 3C-based techniques for analyzing three-dimensional chromatin structure dynamics has driven significant interest in computational methods for 3D chromatin reconstruction. In particular, models based on Hi-C and its single-cell variants, such as scHi-C, have gained widespread popularity. Current approaches for reconstructing the chromatin structure from scHi-C data typically operate by processing one scHi-C map at a time, generating a corresponding 3D chromatin structure as output. Here, we introduce an alternative approach to the whole genome 3D chromatin structure reconstruction that builds upon existing methods while incorporating the broader context of dynamic cellular processes, such as the cell cycle or cell maturation. Our approach integrates scHi-C contact data with single-cell trajectory information and is based on applying simultaneous modeling of a number of cells ordered along the progression of a given cellular process. The approach is able to successfully recreate known nuclear structures while simultaneously achieving smooth, continuous changes in chromatin structure throughout the cell cycle trajectory. Although both Hi-C-based chromatin reconstruction and cellular trajectory inference are well-developed fields, little effort has been made to bridge the gap between them. To address this, we present ChromMovie, a comprehensive molecular dynamics framework for modeling 3D chromatin structure changes in the context of cellular trajectories. To our knowledge, no existing method effectively leverages both the variability of single-cell Hi-C data and explicit information from estimated cellular trajectories, such as cell cycle progression, to improve chromatin structure reconstruction.
Keywords: Chromatin Structure; Molecular Dynamics; Single-Cell Hi-C; Trajectory Inference.
Conflict of interest statement
Competing interests No competing interest is declared.
Figures



References
-
- Jerkovic Ivana and Cavalli Giacomo. Understanding 3d genome organization by multidisciplinary methods. Nature Reviews Molecular Cell Biology, 22:511–528, 2021. - PubMed
-
- Cremer T, Cremer C, Schneider T, Baumann H, Hens L, and Kirsch-Volders M. Analysis of chromosome positions in the interphase nucleus of chinese hamster cells by laseruv-microirradiation experiments. Hum Genet., 62:201–209, 1982. - PubMed
-
- Trzaskoma Paweł, Ruszczycki Błażej, Lee Byoungkoo, Pels Katarzyna K., Krawczyk Katarzyna, Bokota Grzegorz, Szczepankiewicz Andrzej A., Aaron Jesse, Walczak Agnieszka, Ma lgorzata A. Śliwińska , Adriana Magałska Michal Kadlof, Wolny Artur, Parteka Zofia, Arabasz Sebastian, Magdalena Kiss-Arabasz Dariusz Plewczynski, Ruan Yijun, and Wilczyński Grzegorz M.. Ultrastructural visualization of 3d chromatin folding using volume electron microscopy and dna in situ hybridization. Nature Communications, 11, 2020. - PMC - PubMed
-
- Shtengel Gleb, Galbraith James A., Galbraith Catherine G., Jennifer Lippincott-Schwartz Jennifer M. Gillette, Manley Suliana, Sougrat Rachid, Waterman Clare M., Kanchanawong Pakorn, Davidson Michael W., Fetter Richard D., and Hess Harald F.. Interferometric fluorescent super-resolution microscopy resolves 3d cellular ultrastructure. Proceedings of the National Academy of Sciences, 106(9):3125–3130, 2009. - PMC - PubMed
-
- Kadlof Michał, Banecki Krzysztof, Chiliński Mateusz, and Plewczynski Dariusz. Chromatin image-driven modelling. Methods, 226:54–60, 2024. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
