This is a preprint.
Phosphorylation enables allosteric control of a viral condensate
- PMID: 40475503
- PMCID: PMC12139845
- DOI: 10.1101/2025.05.24.655949
Phosphorylation enables allosteric control of a viral condensate
Abstract
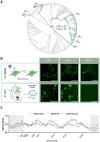
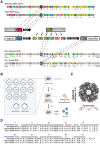
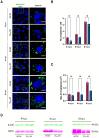
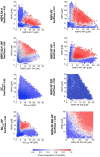
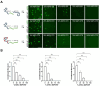
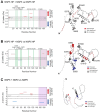
In many viruses, intrinsically disordered proteins (IDPs) drive the formation of replicative organelles essential for viral production. In species A rotaviruses, the disordered protein NSP5 forms condensates in cells via liquid-liquid phase separation (LLPS). Yet the sequence diversity of NSP5 raises the question of whether condensate formation is conserved across all strains and if distinct variants employ alternative mechanisms for nucleating phase separation. Using a machine learning approach, we demonstrate that NSP5 variants differ significantly in their propensity to phase-separate. We engineered a variant incorporating amino acid signatures from strains with low LLPS tendency, which failed to phase separate in vitro yet supported the formation of replicative condensates in recombinant viruses in cells. Low-tendency LLPS strains require phosphorylation of NSP5 to nucleate phase separation, whereas high-tendency strains do not, suggesting distinct nucleation mechanisms. Furthermore, hydrogen-deuterium exchange mass spectrometry revealed a phosphorylation-driven allosteric switch between binding sites on the high-propensity variant. These findings establish that phosphorylation plays a context-dependent role in the formation of replicative organelles across diverse rotaviruses.
Conflict of interest statement
Conflict of interest The authors declare no conflict of interest.
Figures









References
-
- Arosio P., Müller T., Rajah L., Yates E.V., Aprile F.A., Zhang Y., Cohen S.I.A., White D.A., Herling T.W., De Genst E.J., Linse S., Vendruscolo M., Dobson C.M., Knowles T.P.J., 2016. Microfluidic Diffusion Analysis of the Sizes and Interactions of Proteins under Native Solution Conditions. ACS Nano 10, 333–341. 10.1021/acsnano.5b04713 - DOI - PubMed
-
- Arter W.E., Qi R., Erkamp N.A., Krainer G., Didi K., Welsh T.J., Acker J., Nixon-Abell J., Qamar S., Guillén-Boixet J., Franzmann T.M., Kuster D., Hyman A.A., Borodavka A., George-Hyslop P.S., Alberti S., Knowles T.P.J., 2022. Biomolecular condensate phase diagrams with a combinatorial microdroplet platform. Nat Commun 13, 7845. 10.1038/s41467-022-35265-7 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
