This is a preprint.
A Universal 6iL/E4 Culture System for Deriving and Maintaining Embryonic Stem Cells Across Mammalian Species
- PMID: 40475639
- PMCID: PMC12139945
- DOI: 10.1101/2025.05.20.654948
A Universal 6iL/E4 Culture System for Deriving and Maintaining Embryonic Stem Cells Across Mammalian Species
Abstract
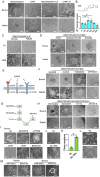
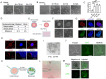
The derivation of authentic embryonic stem cells (ESCs) from diverse mammalian species offers valuable opportunities for advancing regenerative medicine, studying developmental biology, and enabling species conservation. Here, we report the development of a robust, serum-free culture system, termed 6iL/E4 that enables the derivation and long-term self-renewal of ESCs from multiple mammalian species, including mouse, rat, bovine, rabbit, and human. Using systematic signaling pathway analysis, we identified key regulators-including GSK3α, STAT3, PDGFR, BRAF, and LATS-critical for ESC maintenance across species. Additionally, inducible expression of KLF2 and NANOG enhances the naive pluripotency and chimeric potential of bovine ESCs. The E4 medium also supports stable ESC growth while minimizing lineage bias. These findings reveal conserved principles underlying ESC self-renewal across divergent mammalian species and provide a universal platform for cross-species stem cell research, disease modeling, and biotechnology applications.
Keywords: 6iL/E4 culture; GSK3α; PDGFR signaling; bovine ESCs; chimerism; embryonic stem cells; naive pluripotency; pluripotent stem cells; rabbit ESCs; species conservation.
Conflict of interest statement
DECLARATION OF INTERESTS Y.E.C., X.X., and J.X. are equity holders of ATGC Inc. Three provisional patents related to this study have been filed (APPLICATION # 63/798,735; APPLICATION # 63/798,645; APPLICATION # 63/748,241).
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
