Long-term Metformin Alters Gut Microbiota and Serum Metabolome in Coronary Artery Disease Patients After Percutaneous Coronary Intervention to Improve 5-year Prognoses: A Multi-omics Analysis
- PMID: 40475712
- PMCID: PMC12135650
- DOI: 10.31083/RCM26835
Long-term Metformin Alters Gut Microbiota and Serum Metabolome in Coronary Artery Disease Patients After Percutaneous Coronary Intervention to Improve 5-year Prognoses: A Multi-omics Analysis
Abstract
Background: About 20% of patients with coronary artery disease (CAD) experience adverse events within five years of undergoing percutaneous coronary intervention (PCI) for acute myocardial infarction. In these patients, the impact of metformin on long-term prognosis remains uncertain.
Methods: This study enrolled 22 metformin (Met)-CAD patients with diabetes mellitus (DM) who had been administered metformin for at least six months before PCI, 14 non-Met CAD-DM patients with DM who had never taken metformin or had stopped taking metformin for a year before PCI, and 22 matched healthy controls. A 5-year follow-up was conducted to collect clinical prognosis data. Fecal 16S rRNA sequencing and serum untargeted metabolomics analyses were performed. BugBase was utilized to analyze the possible functional changes in the gut microbiome. Multi-omics analysis was conducted using Spearman's correlation to explore the interactions between metformin, gut microbiome, serum metabolites, and clinical prognosis.
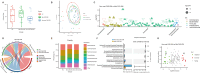
Results: Metformin significantly lowered the 5-year major adverse cardiac events (MACEs) in Met CAD-DM patients. We found a higher abundance of Bacteroides coprocola, Bacteroides massiliensis, Phascolarctobacterium succinatutens, and Eubacterium coprostanoligenes in the Met CAD-DM patients, as well as an increase in hydroxy-alpha-sanshool (HAS) and decenoylcarnitine and a decrease in tridec-10-enoic acid, Z-vad-fmk (benzyloxycarbonyl-Val-Ala-Asp (OMe)-fluoromethylketone), 3,9-dimethyluric acid in blood serum. Multi-omics analysis revealed that alterations in the gut microbiome and serum metabolites are significantly associated with the 5-year prognosis of CAD-DM.
Conclusions: Metformin significantly improved the 5-year prognosis of CAD patients following PCI. Metformin tended to have more positive effects on the commensal flora and metabolic profiles, which may explain its beneficial effects on cardiovascular health. This study revealed the potential associations between metformin and the gut microbiome, an associated alteration in serum metabolome, and the impact on the host immune system and metabolic pathways.
Keywords: coronary artery disease; diabetes mellitus; gut microbiota; metformin; multiomic analyses.
Copyright: © 2025 The Author(s). Published by IMR Press.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Stachon P, Kaier K, Hehn P, Peikert A, Wolf D, Oettinger V, et al. Coronary artery bypass grafting versus stent implantation in patients with chronic coronary syndrome and left main disease: insights from a register throughout Germany. Clinical Research in Cardiology: Official Journal of the German Cardiac Society . 2022;111:742–749. doi: 10.1007/s00392-021-01931-x. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

