Comparative transcriptome analysis reveals key long noncoding RNAs for cadmium tolerance in Tibetan hull-less barley
- PMID: 40475905
- PMCID: PMC12138524
- DOI: 10.3389/fpls.2025.1572490
Comparative transcriptome analysis reveals key long noncoding RNAs for cadmium tolerance in Tibetan hull-less barley
Abstract
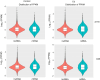
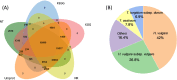
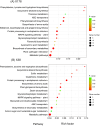
Cadmium (Cd) is one of the most hazardous and persistent heavy metal pollutants globally. Long noncoding RNAs (lncRNAs) play a crucial role in regulating plant gene expression under various abiotic stress conditions. This study investigated the response of the lncRNA transcriptome in the roots of two contrasting Tibetan hull-less barley genotypes, X178 (Cd-tolerant) and X38 (Cd-sensitive), to Cd stress using RNA sequencing. A total of 8299 novel lncRNAs were identified, with 5166 unique target genes associated with 2571 unique lncRNAs. Among these, 1884 target genes were regulated by cis-acting lncRNAs, while 3428 were regulated by trans-acting lncRNAs. By analyzing differential expression profiles in the two genotypes under Cd stress, 26 lncRNAs and 150 mRNAs were identified as potentially linked to Cd tolerance. Functional enrichment analysis revealed that the target genes were significantly enriched in detoxification and stress response functions, including pathways related to phenylalanine, tyrosine, tryptophan, ABC transporters, and secondary metabolites. Additionally, 12 lncRNAs forming 18 lncRNA-mRNA pairs were identified as key regulators of Cd tolerance. The functional roles of these lncRNA-mRNA interactions suggest that they modulate proteins such as DJ-1, EDR, PHT, and ABC transporters, which may contribute to the Cd tolerance observed in genotype X178. High-throughput sequencing results were validated by qRT-PCR. These findings deepen our understanding of lncRNAs as critical regulators of Cd tolerance in plants, offering valuable insights into the molecular mechanisms underlying heavy metal stress responses in crops.
Keywords: Cd toxicity; Hordeum vulgare var. nudum; high-throughput sequencing; lncRNA; mRNA; target genes.
Copyright © 2025 Foysal, Qiu and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Comprehensive Physio-Biochemical Evaluation Reveals Promising Genotypes and Mechanisms for Cadmium Tolerance in Tibetan Hull-Less Barley.Plants (Basel). 2024 Dec 23;13(24):3593. doi: 10.3390/plants13243593. Plants (Basel). 2024. PMID: 39771291 Free PMC article.
-
Comparative Long Non-Coding Transcriptome Analysis of Three Contrasting Barley Varieties in Response to Aluminum Stress.Int J Mol Sci. 2024 Aug 23;25(17):9181. doi: 10.3390/ijms25179181. Int J Mol Sci. 2024. PMID: 39273130 Free PMC article.
-
Uncovering the key lncRNAs in regulating cadmium accumulation and translocation in sweet sorghum.Planta. 2024 Dec 11;261(1):12. doi: 10.1007/s00425-024-04589-7. Planta. 2024. PMID: 39661199
-
Genome-wide transcriptome and functional analysis of two contrasting genotypes reveals key genes for cadmium tolerance in barley.BMC Genomics. 2014 Jul 19;15(1):611. doi: 10.1186/1471-2164-15-611. BMC Genomics. 2014. PMID: 25038590 Free PMC article.
-
Long non-coding RNAs: emerging players regulating plant abiotic stress response and adaptation.BMC Plant Biol. 2020 Oct 12;20(1):466. doi: 10.1186/s12870-020-02595-x. BMC Plant Biol. 2020. PMID: 33046001 Free PMC article. Review.
References
-
- Bai Y., He J., Yao Y., An L., Cui Y., Li X., et al. . (2024). Identification and functional analysis of long non-coding RNA (lncRNA) and metabolites response to mowing in hulless barley (Hordeum vulgare L. var. nudum hook. f.). BMC Plant Biol. 24, 1–15. doi: 10.1186/s12870-024-05334-8 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

