Organoids technology in cancer research: from basic applications to advanced ex vivo models
- PMID: 40476002
- PMCID: PMC12137298
- DOI: 10.3389/fcell.2025.1569337
Organoids technology in cancer research: from basic applications to advanced ex vivo models
Abstract
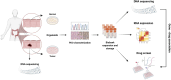
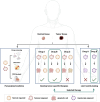
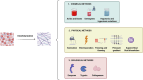
Patient-derived organoids (PDOs) are tridimensional cultures derived from the stem component of a tissue. They preserve the genetic and phenotypic characteristics of the tissue of origin, and represent valuable in vitro models for drug screening, biomarker discovery, cell therapy and genetic modification. Importantly, PDOs reproduce the tumor behavior and can predict therapeutic responses, making them relevant for clinical applications for personalized therapies. PDOs may also be used for studying the interactions between cancer cells and the tumor microenvironment (TME). These interactions are driven by biochemical factors released by the cells, and biomechanical events such as the remodeling of the extracellular matrix (ECM). In recent years, it has become evident that the interactions between cancer cells and the TME have an impact on tumor development and on the efficacy of cancer therapy Therefore, targeting both tumor cells and the TME may improve patient response to treatment. Most PDO culture protocols are limited to epithelial cells. However, recent advances such as use of decellularized ECM (dECM) scaffolds have allowed for the development of in vivo-like environments that host diverse cell types, both normal and pathological, in a tridimensional (3D) manner that closely mimics the complexity of the TME. dECM-based models effectively replicate the interactions between tumor cells, ECM and the microenvironment, are easy to analyze and adaptable for drug testing. By incorporating TME components and therapeutic agents, these models offer an advanced platform for preclinical testing.
Keywords: cancer organoids; decellularized matrix; drug screening; ex vivo cancer models; extracellular matrix (ECM); personalized therapy.
Copyright © 2025 Varinelli, Illescas, Lorenc, Battistessa, Di Bella, Zanutto and Gariboldi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Patient Derived Organoids (PDOs), Extracellular Matrix (ECM), Tumor Microenvironment (TME) and Drug Screening: State of the Art and Clinical Implications of Ovarian Cancer Organoids in the Era of Precision Medicine.Cancers (Basel). 2023 Mar 30;15(7):2059. doi: 10.3390/cancers15072059. Cancers (Basel). 2023. PMID: 37046719 Free PMC article. Review.
-
Patient derived organoids in prostate cancer: improving therapeutic efficacy in precision medicine.Mol Cancer. 2021 Sep 29;20(1):125. doi: 10.1186/s12943-021-01426-3. Mol Cancer. 2021. PMID: 34587953 Free PMC article. Review.
-
Role of the tumor microenvironment in malignant melanoma organoids during the development and metastasis of tumors.Front Cell Dev Biol. 2023 Apr 19;11:1166916. doi: 10.3389/fcell.2023.1166916. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37152280 Free PMC article. Review.
-
Patient-derived organoids in precision cancer medicine.Med. 2024 Nov 8;5(11):1351-1377. doi: 10.1016/j.medj.2024.08.010. Epub 2024 Sep 27. Med. 2024. PMID: 39341206 Review.
-
Decellularized extracellular matrix-decorated 3D nanofiber scaffolds enhance cellular responses and tissue regeneration.Acta Biomater. 2024 Aug;184:81-97. doi: 10.1016/j.actbio.2024.06.020. Epub 2024 Jun 21. Acta Biomater. 2024. PMID: 38908416
Cited by
-
Decoding the role of cancer stem cells in digestive tract tumors: Mechanisms and therapeutic implications (Review).Int J Oncol. 2025 Jul;67(1):61. doi: 10.3892/ijo.2025.5767. Epub 2025 Jun 27. Int J Oncol. 2025. PMID: 40576138 Free PMC article. Review.
References
-
- Alessandrini A., Facci P. (2005). AFM: a versatile tool in biophysics. Meas. Sci. Technol. 16, R65–R92. 10.1088/0957-0233/16/6/R01 - DOI
Publication types
LinkOut - more resources
Full Text Sources

