Molecular and morphological data support the synonymy of Muricanthusradix Gmelin, 1791 and Muricanthusambiguus Reeve, 1845 (Gastropoda, Muricidae)
- PMID: 40476220
- PMCID: PMC12138812
- DOI: 10.3897/zookeys.1239.143837
Molecular and morphological data support the synonymy of Muricanthusradix Gmelin, 1791 and Muricanthusambiguus Reeve, 1845 (Gastropoda, Muricidae)
Abstract
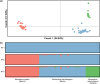
The Muricanthusradix/ambiguus/nigritus complex includes species with a great diversity of shell shapes and shared habitats in various regions, which has raised questions and doubts about the current taxonomic classification of these species. Muricanthusnigritus, M.radix, and M.ambiguus are three similar-looking black and white murex found commonly on the west coast of North and South America. The wide variety of morphological patterns within and between these species makes the classification of specimens difficult by visual observation. To this day, controversy persists over whether M.radix and M.ambiguus are one or two distinct species. Molecular genetic data have helped clarify the taxonomic classification of many mollusk species in recent decades, contributing to a more accurate understanding of biodiversity and ecosystems. In this study, DNA barcoding and double digest restriction-site associated DNA sequencing (ddRAD-seq) methodologies were applied to complement morphological data, establishing for the first time the phylogenetic relationships between M.nigritus, M.ambiguus and M.radix. The classic mitochondrial and nuclear barcodes obtained from 80 specimens collected from three different geographic locations differentiated only two phylogenetic clades (M.nigritus and M.radix/ambiguus from Mexico differentiated from M.radix/ambiguus from Mexico and Panama). High levels of mitochondrial DNA introgression have been observed between M.nigritus and M.radix/ambiguus. The deep-level approach performed using 3692 loci obtained from ddRAD-seq also differentiated only two genetic clusters (M.nigritus and M.radix/ambiguus). Our results clearly support the proposal that M.ambiguus should be synonymized with M.radix.
Keywords: Muricanthus; ddRADseq; mithocondrial genes; morphometrics; nuclear genes; phylogeny; taxonomy.
Francisco Morinha, James Ernest, Ana Archer-Taveira, Ana M. Rocha, Robert T. Iwamasa.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Hydroids (Cnidaria, Hydrozoa) from Mauritanian Coral Mounds.Zootaxa. 2020 Nov 16;4878(3):zootaxa.4878.3.2. doi: 10.11646/zootaxa.4878.3.2. Zootaxa. 2020. PMID: 33311142
-
Genome-wide SNP marker discovery and phylogenetic analysis of mulberry varieties using double-digest restriction site-associated DNA sequencing.Gene. 2020 Feb 5;726:144162. doi: 10.1016/j.gene.2019.144162. Epub 2019 Oct 19. Gene. 2020. PMID: 31639429
-
Scrutinising an inscrutable bark-nesting ant: Exploring cryptic diversity in the Rhopalomastix javana (Hymenoptera: Formicidae) complex using DNA barcodes, genome-wide MIG-seq and geometric morphometrics.PeerJ. 2023 Nov 16;11:e16416. doi: 10.7717/peerj.16416. eCollection 2023. PeerJ. 2023. PMID: 38025712 Free PMC article.
-
European Lymnaeidae (Mollusca: Gastropoda), intermediate hosts of trematodiases, based on nuclear ribosomal DNA ITS-2 sequences.Infect Genet Evol. 2001 Dec;1(2):85-107. doi: 10.1016/s1567-1348(01)00019-3. Infect Genet Evol. 2001. PMID: 12798024
-
Unravelling the riddle of Radix: DNA barcoding for species identification of freshwater snail intermediate hosts of zoonotic digeneans and estimating their inter-population evolutionary relationships.Infect Genet Evol. 2015 Oct;35:63-74. doi: 10.1016/j.meegid.2015.07.021. Epub 2015 Jul 18. Infect Genet Evol. 2015. PMID: 26196736
References
-
- Barco A, Claremont M, Reid DG, Houart R, Bouchet P, Williams ST, Cruaud C, Couloux A, Oliverio M. (2010) A molecular phylogenetic framework for the Muricidae, a diverse family of carnivorous gastropods. Molecular Phylogenetics and Evolution 56(3): 1025–1039. 10.1016/j.ympev.2010.03.008 - DOI - PubMed
-
- Borges LM, Hollatz C, Lobo J, Cunha AM, Vilela AP, Calado G, Coelho R, Costa AC, Ferreira MSG, Costa MH, Costa FO. (2016) With a little help from DNA barcoding: Investigating the diversity of Gastropoda from the Portuguese coast. Scientific Reports 6(1): 20226. 10.1038/srep20226 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
