A reference genome for Trichogramma kaykai: a tiny desert-dwelling parasitoid wasp with competing sex-ratio distorters
- PMID: 40476364
- PMCID: PMC12341880
- DOI: 10.1093/g3journal/jkaf129
A reference genome for Trichogramma kaykai: a tiny desert-dwelling parasitoid wasp with competing sex-ratio distorters
Abstract
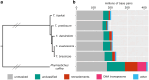
The tiny parasitoid wasp Trichogramma kaykai inhabits the Mojave Desert of the southwest United States. Populations of this tiny insect variably host up to 2 different sex-distorting genetic elements: (1) the endosymbiotic bacterium Wolbachia which induces the parthenogenetic reproduction of females, and (2) a B-chromosome, "Paternal Sex Ratio" (PSR), which converts would-be female offspring to PSR-transmitting males. We report here the genome of a Wolbachia-infected T. kaykai isofemale colony KSX58. Using Oxford Nanopore sequencing, we produced a final genome assembly of 205 Mbp with 34× coverage, consisting of 154 contigs with an N50 of 2.2 Mbp. The assembly is quite complete, with 92.67% complete Hymenoptera BUSCOs recovered: a very high score for Trichogrammatids that have been previously characterized for having high levels of core gene losses. We also report a complete mitochondrial genome for T. kaykai, and an assembly of the associated Wolbachia, strain wTkk. Finally, we identified copies of the parthenogenesis-inducing (PI) genes pifA and pifB in a remnant prophage region of the wTkk genome and compared their evolution to pifs from a suite of other PI Wolbachia. The T. kaykai assembly is one of the highest quality genome assemblies for the genus to date and will serve as a great resource for understanding the evolution of sex and selfish genetic elements.
Keywords: Wolbachia; B-chromosome; genome assembly; parthenogenesis; selfish genetic element; sex ratio; symbiosis.
© The Author(s) 2025. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The authors declare no conflict of interest.
Figures



Update of
-
A reference genome for Trichogramma kaykai: A tiny desert-dwelling parasitoid wasp with competing sex-ratio distorters.bioRxiv [Preprint]. 2024 Nov 23:2024.11.22.624848. doi: 10.1101/2024.11.22.624848. bioRxiv. 2024. Update in: G3 (Bethesda). 2025 Aug 6;15(8):jkaf129. doi: 10.1093/g3journal/jkaf129. PMID: 39605481 Free PMC article. Updated. Preprint.
Similar articles
-
A reference genome for Trichogramma kaykai: A tiny desert-dwelling parasitoid wasp with competing sex-ratio distorters.bioRxiv [Preprint]. 2024 Nov 23:2024.11.22.624848. doi: 10.1101/2024.11.22.624848. bioRxiv. 2024. Update in: G3 (Bethesda). 2025 Aug 6;15(8):jkaf129. doi: 10.1093/g3journal/jkaf129. PMID: 39605481 Free PMC article. Updated. Preprint.
-
Wolbachia symbionts control sex in a parasitoid wasp using a horizontally acquired gene.Curr Biol. 2024 Jun 3;34(11):2359-2372.e9. doi: 10.1016/j.cub.2024.04.035. Epub 2024 Apr 30. Curr Biol. 2024. PMID: 38692276
-
Selfish element maintains sex in natural populations of a parasitoid wasp.Proc Biol Sci. 2001 Mar 22;268(1467):617-22. doi: 10.1098/rspb.2000.1404. Proc Biol Sci. 2001. PMID: 11297179 Free PMC article.
-
Laser therapy for retinopathy in sickle cell disease.Cochrane Database Syst Rev. 2022 Dec 12;12(12):CD010790. doi: 10.1002/14651858.CD010790.pub3. Cochrane Database Syst Rev. 2022. PMID: 36508693 Free PMC article.
-
Dressings and topical agents for treating pressure ulcers.Cochrane Database Syst Rev. 2017 Jun 22;6(6):CD011947. doi: 10.1002/14651858.CD011947.pub2. Cochrane Database Syst Rev. 2017. PMID: 28639707 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
