Genome Structural Variants Shape Adaptive Success of an Invasive Urban Malaria Vector Anopheles stephensi
- PMID: 40476664
- PMCID: PMC12198770
- DOI: 10.1093/molbev/msaf140
Genome Structural Variants Shape Adaptive Success of an Invasive Urban Malaria Vector Anopheles stephensi
Abstract
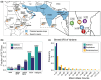
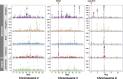
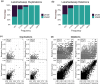
Global changes are associated with the emergence of several invasive species, although genetic determinants of their adaptive success remain poorly understood. To address this problem, we investigated the role genome structural variants (SVs) play in adaptations of Anopheles stephensi, a primary vector of urban malaria in South Asia and an invasive malaria vector in South Asian islands and Africa. Using whole genome sequencing data, we identified 2,988 duplications and 16,038 deletions of SVs in 115 mosquitoes from invasive island populations and four locations from mainland India, the species' ancestral range. The minor allele frequency of SVs and amino acid polymorphism suggests SVs are more deleterious than the amino acid variants. However, high-frequency SVs are enriched in genomic regions with signatures of selective sweeps, implying a putative adaptive role of some SVs. We revealed three novel candidate duplication mutations for recurrent evolution of resistance to diverse insecticides in An. stephensi populations. These mutations exhibit distinct population genetic signatures of recent adaptive evolution, suggesting different mechanisms of rapid adaptations involving hard and soft sweeps helping the species thwart chemical control strategies. We also identify candidate SVs for the larval tolerance to brackish water, which is likely an adaptation in island and coastal populations. Nearly all high-frequency SVs and the candidate adaptive variants in the island populations are derived from the mainland, suggesting a sizable contribution of existing variation to the success of the island populations. Our results highlight the important role of SVs in the evolutionary success of invasive malaria vector An. stephensi.
Keywords: genome structural variation; insecticide resistance; invasive species; population genetics; selective sweep.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest: The authors declare no conflicts of interest.
Figures

Update of
-
Genome structural variants shape adaptive success of an invasive urban malaria vector Anopheles stephensi.bioRxiv [Preprint]. 2024 Jul 30:2024.07.29.605641. doi: 10.1101/2024.07.29.605641. bioRxiv. 2024. Update in: Mol Biol Evol. 2025 Jun 4;42(6):msaf140. doi: 10.1093/molbev/msaf140. PMID: 39211149 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

