Maize big embryo 6 reveals roles of plastidial and cytosolic prephenate aminotransferases in seed and plant development
- PMID: 40476681
- PMCID: PMC12142466
- DOI: 10.1093/plcell/koaf067
Maize big embryo 6 reveals roles of plastidial and cytosolic prephenate aminotransferases in seed and plant development
Abstract
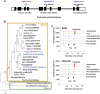
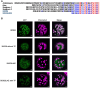
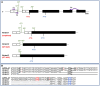
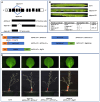
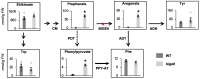
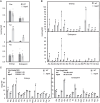
In plants, embryo size is determined via interactions between metabolic and developmental signals. Maize (Zea mays) big embryo 6 (bige6) enhances embryo size while sharply reducing plant growth. Here, we show that BigE6 encodes a plastidial prephenate aminotransferase (PPA-AT), a key enzyme in the arogenate pathway for L-phenylalanine (Phe) and L-tyrosine (Tyr) biosynthesis. The maize BigE6 paralog, BigE6Like, encodes a cytosol-localized PPA-AT, revealing Phe and Tyr biosynthesis via cytosolic arogenate as a potential alternative to the known cytosolic phenylpyruvate pathway. Moreover, the single PPA-AT gene of Arabidopsis (Arabidopsis thaliana) encodes plastidial and cytosolic enzymes by alternative splicing. Transgenic rescue of a ppa-at mutant in Arabidopsis demonstrates that the plastidial PPA-AT is indispensable for seed formation due, in part, to its essential role in the female gametophyte. Leaves of bige6 maize maintained overall homeostasis for aromatic amino acids and downstream metabolites, revealing a resilience of mechanisms that scale growth to a limiting supply of Phe and Tyr. In bige6 seeds, broad perturbation of amino acid homeostasis is associated with transcriptomic upregulation of growth processes in the embryo and endosperm, implicating amino acid signaling in the regulation of embryo size. Our findings reveal the complexity and developmental dependence of growth responses to limiting amino acid biosynthesis.
© The Author(s) 2025. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. There are none to declare.
Figures




Similar articles
-
Phylobiochemical characterization of class-Ib aspartate/prephenate aminotransferases reveals evolution of the plant arogenate phenylalanine pathway.Plant Cell. 2014 Jul;26(7):3101-14. doi: 10.1105/tpc.114.127407. Epub 2014 Jul 28. Plant Cell. 2014. PMID: 25070637 Free PMC article.
-
An alternative pathway contributes to phenylalanine biosynthesis in plants via a cytosolic tyrosine:phenylpyruvate aminotransferase.Nat Commun. 2013;4:2833. doi: 10.1038/ncomms3833. Nat Commun. 2013. PMID: 24270997
-
Completion of the cytosolic post-chorismate phenylalanine biosynthetic pathway in plants.Nat Commun. 2019 Jan 3;10(1):15. doi: 10.1038/s41467-018-07969-2. Nat Commun. 2019. PMID: 30604768 Free PMC article.
-
Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants.J Exp Bot. 2014 Oct;65(19):5527-34. doi: 10.1093/jxb/eru240. Epub 2014 Jun 5. J Exp Bot. 2014. PMID: 24902885 Review.
-
The regulation of zein biosynthesis in maize endosperm.Theor Appl Genet. 2020 May;133(5):1443-1453. doi: 10.1007/s00122-019-03520-z. Epub 2020 Jan 2. Theor Appl Genet. 2020. PMID: 31897513 Review.
Cited by
-
Forever green: A temporal physiologic and metabolic analysis reveals genetic drivers of the staygreen trait in maize.Plant Cell. 2025 Aug 4;37(8):koaf181. doi: 10.1093/plcell/koaf181. Plant Cell. 2025. PMID: 40701651 Free PMC article. No abstract available.
References
-
- Block A, Fristedt R, Rogers S, Kumar J, Barnes B, Barnes J, Elowsky CG, Wamboldt Y, Mackenzie SA, Redding K, et al. Functional modeling identifies paralogous solanesyl-diphosphate synthases that assemble the side chain of plastoquinone-9 in plastids. J Biol Chem. 2013:288(38):27594–27606. 10.1074/jbc.M113.492769 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

