The saga to monitor and control norovirus: the rise of GII.17
- PMID: 40476850
- PMCID: PMC12282288
- DOI: 10.1099/jgv.0.002118
The saga to monitor and control norovirus: the rise of GII.17
Abstract
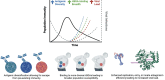
Norovirus is a major cause of acute gastroenteritis in all age groups, with recent surges of cases in Europe and the USA reinforcing the influence of this virus on human health. Despite its societal impact, no vaccine or antiviral drug is available. The development of these countermeasures has been impaired at least in part by the extreme genetic and antigenic diversity of noroviruses. Here, we reviewed historical norovirus outbreaks, including the pandemics of GII.4 norovirus that were first documented in the mid-1990s, sporadic increases of non-GII.4 norovirus (e.g. GII.17 and GII.2) during the 2010s and, most recently, the ongoing large outbreaks caused by a new cluster of GII.17 noroviruses. This five-decade-long journey of tracking noroviruses in the human population illustrates the importance and challenges of battling this evolving virus.
Keywords: antigenic diversity; diarrhea; histo-blood group antigen (HBGA); norovirus; virus evolution.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- Zahorsky J. Hyperemesis hiemis or the winter vomiting disease. Arch Pediatr. 1929;46:391–395.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

