CEBIT screening for inhibitors of the interaction between SARS-CoV-2 spike and ACE2
- PMID: 40477955
- PMCID: PMC8891120
- DOI: 10.1016/j.fmre.2022.01.034
CEBIT screening for inhibitors of the interaction between SARS-CoV-2 spike and ACE2
Abstract
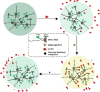
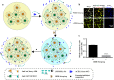
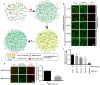
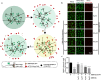
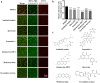
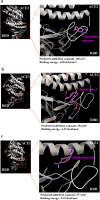
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, causing COVID-19, is the most challenging pandemic of the modern era. It has resulted in over 5 million deaths worldwide. To quickly explore therapeutics for COVID-19, we utilized a previously-established system, namely CEBIT. We performed a high-throughput screening of FDA-approved drugs to inhibit the interaction between the receptor-binding domain (RBD) of SARS-CoV-2 spike protein and its obligate receptor ACE2. This interaction is essential for viral entry and therefore represents a promising therapeutic target. Based on the recruitment of interacting molecules into phase-separated condensates as a readout, we identified six positive candidates from a library of 2572 compounds, most of which have been reported to inhibit the entry of SARS-CoV-2 into host cells. Our surface plasmon resonance (SPR) and molecular docking analyses revealed the possible mechanisms via which these compounds interfere with the interaction between RBD and ACE2. Hence, our results indicate that CEBIT is highly versatile for identifying drugs against SARS-CoV-2 entry, and targeting CoV-2 entry by small molecule drugs is a viable therapeutic option to treat COVID-19 in addition to commonly used monoclonal antibodies.
Keywords: ACE2; High-throughput screening; Protein-protein interaction; SARS-CoV-2; Spike protein.
© 2022 The Authors. Publishing Services by Elsevier B.V. on behalf of KeAi Communications Co. Ltd.
Conflict of interest statement
The authors declare that they have no conflicts of interest in this work.
Figures

References
-
- WHO Coronavirus (COVID-19) Dashboard. Available from: https://covid19.who.int/ (accessed Date 2021).
-
- Hopkins A.L., Groom C.R. The druggable genome. Nat. Rev. Drug Discov. 2002;1(9):727–730. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

