Design and Validation of Hybrid Polymer-Lipid Nanoparticles as Novel Transfection Vectors for MicroRNA Delivery to Human Cardiac Fibroblasts
- PMID: 40478595
- PMCID: PMC12264849
- DOI: 10.1002/adhm.202500971
Design and Validation of Hybrid Polymer-Lipid Nanoparticles as Novel Transfection Vectors for MicroRNA Delivery to Human Cardiac Fibroblasts
Abstract
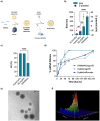
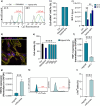
Hybrid polymer-lipid nanoparticles (hybrid NPs) are developed as novel in vitro transfection vectors for microRNAs (miRNAs) delivery to overcome the poor stability, incomplete loading efficiency and fast release kinetics of commercial transfection agents. Hybrid NPs with nanometric size are prepared by a scalable high-yield nanoprecipitation method. They consisted of a lipoplex core, composed of the cationic lipid [2-(2,3-didodecyloxypropyl)-hydroxyethyl] ammonium bromide (DE) and helper lipid dioleoyl phosphatidylethanolamine (DOPE), providing 99% miRNA loading, and a poly(lactic acid-co-glycolic acid) (PLGA) shell, ensuring NPs colloidal stability and controlled miRNA release kinetics. Adult human cardiac fibroblasts (AHCFs), transiently transfected with miR-1 loaded hybrid NPs versus RNAiMAX showed superior viability and higher miRNA content over 48 h. Hybrid NPs could be stored up to 14 days at -20 °C, upon freeze-drying with trehalose cryoprotectant (12% w/v), regaining their physicochemical and biological properties when redispersed. Hybrid NPs are assessed in a miRcombo model of fibroblast-to-cardiomyocyte reprogramming. At 15 days post-transfection with reprogramming miRNAs (miRcombo: miRs-1, 133, 208 and 499), cardiac troponin T marker expression is significantly increased at gene and protein level. These results pave the way to hybrid NP use as transfection vectors for the in vitro testing of miRNAs targeting AHCFs.
Keywords: human cardiac fibroblasts; hybrid nanoparticles; microRNA; nanoparticles storage; transfection vectors.
© 2025 The Author(s). Advanced Healthcare Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
PLGA-LEC/F127 hybrid nanoparticles loaded with curcumin and their modulatory effect on monocytes.Nanomedicine (Lond). 2024 Jun 20;19(15):1407-1423. doi: 10.1080/17435889.2024.2357530. Epub 2024 Jun 26. Nanomedicine (Lond). 2024. PMID: 38920352 Free PMC article.
-
Lipoplexes for effective in vitro delivery of microRNAs to adult human cardiac fibroblasts for perspective direct cardiac cell reprogramming.Nanomedicine. 2022 Sep;45:102589. doi: 10.1016/j.nano.2022.102589. Epub 2022 Jul 28. Nanomedicine. 2022. PMID: 35908737
-
Surface modified poly(β amino ester)-containing nanoparticles for plasmid DNA delivery.J Control Release. 2012 Nov 28;164(1):41-8. doi: 10.1016/j.jconrel.2012.09.020. Epub 2012 Oct 5. J Control Release. 2012. PMID: 23041278 Free PMC article.
-
Lipid-Polymer Hybrid Nanoparticles as a Smart Drug Delivery System for Peptide/Protein Delivery.Pharmaceutics. 2025 Jun 19;17(6):797. doi: 10.3390/pharmaceutics17060797. Pharmaceutics. 2025. PMID: 40574109 Free PMC article. Review.
-
Withdrawal versus continuation of chronic antipsychotic drugs for behavioural and psychological symptoms in older people with dementia.Cochrane Database Syst Rev. 2013 Mar 28;(3):CD007726. doi: 10.1002/14651858.CD007726.pub2. Cochrane Database Syst Rev. 2013. PMID: 23543555
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

