Cryo-EM structures of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase in the apo state and in the presence of small molecules
- PMID: 40478632
- PMCID: PMC12210188
- DOI: 10.1107/S2053230X25004595
Cryo-EM structures of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase in the apo state and in the presence of small molecules
Abstract
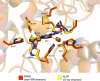
Unlike humans, Mycobacterium tuberculosis (Mtb), the causative agent of human tuberculosis, has a de novo histidine-biosynthesis pathway. The enzyme imidazole glycerol phosphate dehydratase (IGPD), which catalyses the conversion of imidazole glycerol phosphate to imidazole acetol phosphate, has been studied extensively from various organisms and has become a major target for the development of antibacterial, antiweed and antifungal small molecules. In our previous studies, we have shown that in crystals IGPD forms a 24-mer oligomeric state in which the monomers are arranged in 432 symmetry. In order to gain insights into the oligomeric state of Mtb IGPD in solution, we determined cryogenic sample electron microscopy (cryo-EM) structures of apo IGPD at 2.2 and 3.1 Å resolution. In addition, we also determined the cryo-EM structure of IGPD in the presence of 3-amino-1,2,4-triazole (ATZ) to 2.8 Å resolution. The results of this work, which corroborate those from the crystallographic studies, indicate that IGPD forms a homo-oligomeric structure in solution comprising of 24 subunits. ATZ binds in the active-site pocket of the enzyme, which is located at the interface of three monomers and tethers 24 ATZ molecules. The results of this study suggest that cryo-EM, in addition to being a rapidly evolving and complementary imaging technology for elucidating 3D structures of biological macromolecules, can be useful in pinpointing the mode of binding small molecules of low mass (here ∼85 Da) and mapping protein-ligand interactions, which could assist in the design of accurate (high-potency) inhibitors.
Keywords: cryo-EM; drug discovery; histidine-biosynthesis pathway; imidazole glycerol phosphate dehydratase; tuberculosis.
Figures



Similar articles
-
Structures of native, substrate-bound and inhibited forms of Mycobacterium tuberculosis imidazoleglycerol-phosphate dehydratase.Acta Crystallogr D Biol Crystallogr. 2013 Dec;69(Pt 12):2461-7. doi: 10.1107/S0907444913022579. Epub 2013 Nov 19. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 24311587
-
Crystal structures of Mycobacterium tuberculosis and Mycobacterium thermoresistibile glycyl-tRNA synthetases in various liganded states.PLoS One. 2025 Jun 30;20(6):e0326500. doi: 10.1371/journal.pone.0326500. eCollection 2025. PLoS One. 2025. PMID: 40587479 Free PMC article.
-
Structural basis of nearest-neighbor cooperativity in the ring-shaped gene regulatory protein TRAP from protein engineering and cryo-EM.Proc Natl Acad Sci U S A. 2025 Jan 7;122(1):e2409030121. doi: 10.1073/pnas.2409030121. Epub 2024 Dec 30. Proc Natl Acad Sci U S A. 2025. PMID: 39793047 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Ahangar, M. S., Vyas, R., Nasir, N. & Biswal, B. K. (2013). Acta Cryst. D69, 2461–2467. - PubMed
-
- Caminero, J. A., Sotgiu, G., Zumla, A. & Migliori, G. B. (2010). Lancet Infect. Dis.10, 621–629. - PubMed
-
- Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., Tekaia, F., Badcock, K., Basham, D., Brown, D., Chillingworth, T., Connor, R., Davies, R., Devlin, K., Feltwell, T., Gentles, S., Hamlin, N., Holroyd, S., Hornsby, T., Jagels, K., Krogh, A., McLean, J., Moule, S., Murphy, L., Oliver, K., Osborne, J., Quail, M. A., Rajandream, M., Rogers, J., Rutter, S., Seeger, K., Skelton, J., Squares, R., Squares, S., Sulston, J. E., Taylor, K., Whitehead, S. & Barrell, B. G. (1998). Nature, 393, 537–544. - PubMed
MeSH terms
Substances
Grants and funding
- DBT/PR12422/MED/31/287/2014/Department of Biotechnology, Ministry of Science and Technology, India
- BT/PR29075/BRB/10/1699/2018/Department of Biotechnology, Ministry of Science and Technology, India
- BT/PR40325/BTIS/137/1/2020/Department of Biotechnology, Ministry of Science and Technology, India
- BT/HRD/TIF/09/02/2021-22/Department of Biotechnology, Ministry of Science and Technology, India
- RTI 4006/Department of Atomic Energy, Government of India
LinkOut - more resources
Full Text Sources
Miscellaneous

