SARS-COV-2 mutations in North Rift, Kenya
- PMID: 40478857
- PMCID: PMC12143566
- DOI: 10.1371/journal.pone.0325133
SARS-COV-2 mutations in North Rift, Kenya
Abstract
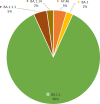
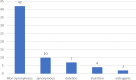
The rise of new SARS-CoV-2 mutations brought challenges and progress in the global fight against COVID-19. Mutations in spike and accessory genes affect transmission, vaccine efficacy, treatments, testing, and public health strategies. Monitoring emerging variants is crucial to halt re-emergency of the virus and spread. 44 nasopharyngeal/oropharyngeal swabs from Kenyan patients were sequenced with the Illumina platform. Galaxy's bioinformatic tools were used for genomic analysis. SARS-CoV-2 genome classification was done using PANGOLIN and mutation annotation with the COVID-19 Annotator tool. From this study, 5 clades of SARS-CoV-2 were identified of whom 38 (86%) were BA.1.1; 2 (5%) were BA.1.1.1; 1 (2%) was BA.1; 1 (2%) was BA.1.14 and 2 (5%) were AY.46. Symptomatic patients were 16 out of 18 males and 22 out of 26 females. Out of these, symptomatic patients, BA.1.1 was found in 14 males and 18 females. In these clades we found 53 significant mutations of which 42 were non-synonymous, 10 synonymous, 7 deletions, 4 insertions and 2 extragenic. Out of the 42 non-synonymous mutations, 7 were exclusively found in symptomatic patients. Two new mutations, S:R214R, and NSP2:A555A, were also found and were dominant in symptomatic patients. These findings add to the understanding of the SARS-CoV-2 virus future evolution in the region.
Copyright: © 2025 Mbogori et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Genomic characterization of SARS-CoV-2 isolates from patients in Turkey reveals the presence of novel mutations in spike and nsp12 proteins.J Med Virol. 2021 Oct;93(10):6016-6026. doi: 10.1002/jmv.27188. Epub 2021 Jul 16. J Med Virol. 2021. PMID: 34241906 Free PMC article.
-
Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19).2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 34033342 Free Books & Documents.
-
COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest.Elife. 2021 Feb 23;10:e63409. doi: 10.7554/eLife.63409. Elife. 2021. PMID: 33620031 Free PMC article.
-
The variants question: What is the problem?J Med Virol. 2021 Dec;93(12):6479-6485. doi: 10.1002/jmv.27196. Epub 2021 Jul 28. J Med Virol. 2021. PMID: 34255352 Free PMC article. Review.
-
Current understanding of the adaptive evolution of the SARS-CoV-2 genome.Yi Chuan. 2025 Feb;47(2):211-227. doi: 10.16288/j.yczz.24-231. Yi Chuan. 2025. PMID: 39924701 Review.
References
-
- Khatri R, Siddqui G, Sadhu S, Maithil V, Vishwakarma P, Lohiya B, et al.. Intrinsic D614G and P681R/H mutations in SARS-CoV-2 VoCs alpha, delta, omicron and viruses with D614G plus key signature mutations in spike protein alters fusogenicity and infectivity. Med Microbiol Immunol. 2023;212(1):103–22. doi: 10.1007/s00430-022-00760-7 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous