Bacterial RecD2 is a processive single-stranded DNA translocase with strand-switching capacity at DNA forks
- PMID: 40479711
- PMCID: PMC12143592
- DOI: 10.1093/nar/gkaf459
Bacterial RecD2 is a processive single-stranded DNA translocase with strand-switching capacity at DNA forks
Abstract
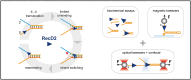
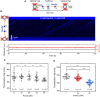
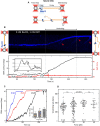
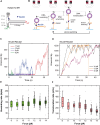
RecD2 is a superfamily 1B helicase involved in DNA replication and repair, modulating replication restart, fork progression, and RecA recombinase activity. In this work, we have characterized the functions of Bacillus subtilis RecD2 using biochemical and single-molecule approaches. ATPγS binding and low MgCl2 concentrations enhance DNA association, with a preference for forked structures and unstructured DNA longer than 30 nucleotides. RecD2 binds to end-less single-stranded DNA stretched at 8-20 pN and translocates through ATP hydrolysis over long distances (>20 kb) with 5'-3' polarity at high rates. RecD2 shows limited unwinding activity on fork structures, strongly dependent on protein concentration and duplex length, reflecting low processivity. However, processivity improves significantly when force is applied to the translocating strand or unwound DNA ends, enabling the unwinding of thousands of base pairs at rates up to 160 bp/s. Single-molecule assays reveal frequent strand switching on fork substrates, resulting in a non-productive cycle of unwinding and rewinding, likely mediated by the N-terminal domain. This behavior explains the low helicase activity observed in bulk assays. We propose that regulation of strand-switching activity may be relevant for RecD2's in vivo function.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

