Unlocking the serine mischarging paradox and inhibiting lactyltransferase activity of AlaRS by a single-point mutation
- PMID: 40479712
- PMCID: PMC12143591
- DOI: 10.1093/nar/gkaf462
Unlocking the serine mischarging paradox and inhibiting lactyltransferase activity of AlaRS by a single-point mutation
Abstract
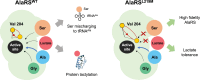
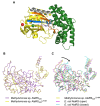
Aminoacyl-tRNA synthetases are critical for accurate genetic translation, attaching amino acids to their corresponding transfer RNA molecules. Alanyl-tRNA synthetase (AlaRS) often misactivates Ser or Gly instead of Ala, which is detrimental unless corrected by its editing functions. The paradox of misactivating larger Ser by AlaRS was considered inevitable due to its inherent design, sharing an essential acidic residue to accommodate the activated adenylated intermediates from both cognate and non-cognate amino acids. Here we show a groundbreaking discovery where a single-point mutation, L219M, in AlaRS from Methylomonas sp. DH-1, effectively eliminates Ser misactivation. Structural analysis of the pre-activation state unveiled that the flexibility of Val204 is the key to preventing Ser binding in AlaRSL219M. This research elucidates the amino acid discrimination mechanism in AlaRS, independent of editing domain. Remarkably, the AlaRSL219M mutation was initially identified as a causal mutation enhancing lactate tolerance in a strain developed through adaptive laboratory evolution. We showed that AlaRSL219M also eliminates the enzyme's inherent lactyltransferase activity, suggesting that the lactate tolerance observed might result from preventing excessive protein lactylation under lactate stress. This opens possibilities for developing high-fidelity and lactylation-deficient AlaRS mutants across various organisms, facilitating studies on their potential benefits in different physiological scenarios.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures




Similar articles
-
Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.Nature. 2009 Dec 10;462(7274):808-12. doi: 10.1038/nature08612. Nature. 2009. PMID: 20010690 Free PMC article.
-
Substrate specificity and catalysis by the editing active site of Alanyl-tRNA synthetase from Escherichia coli.Biochemistry. 2011 Mar 8;50(9):1474-82. doi: 10.1021/bi1013535. Epub 2011 Jan 31. Biochemistry. 2011. PMID: 21241052 Free PMC article.
-
Fine-Tuning of Alanyl-tRNA Synthetase Quality Control Alleviates Global Dysregulation of the Proteome.Genes (Basel). 2020 Oct 18;11(10):1222. doi: 10.3390/genes11101222. Genes (Basel). 2020. PMID: 33081015 Free PMC article.
-
The uniqueness of AlaRS and its human disease connections.RNA Biol. 2021 Nov;18(11):1501-1511. doi: 10.1080/15476286.2020.1861803. Epub 2020 Dec 23. RNA Biol. 2021. PMID: 33317386 Free PMC article. Review.
-
A metal-binding motif implicated in RNA recognition by an aminoacyl-tRNA synthetase and by a retroviral gene product.Mol Microbiol. 1992 May;6(10):1259-62. doi: 10.1111/j.1365-2958.1992.tb00846.x. Mol Microbiol. 1992. PMID: 1379318 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

