Selective engineering of condensation properties of single-stranded DNA binding (SSB) protein via its intrinsically disordered linker region
- PMID: 40479713
- PMCID: PMC12143593
- DOI: 10.1093/nar/gkaf481
Selective engineering of condensation properties of single-stranded DNA binding (SSB) protein via its intrinsically disordered linker region
Abstract
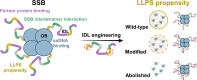
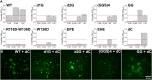
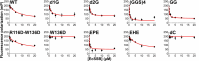
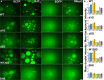
Single-stranded DNA binding (SSB) proteins are essential components of genome metabolism in both bacteria and eukaryotes. Recently demonstrated condensation propensities have placed SSB functions in a new context regarding the organization of nucleic acid-modifying complexes. In this work, we provide functional dissection of the condensation and partner binding properties of Escherichia coli (Ec) SSB via engineered modifications of its intrinsically disordered linker (IDL) region. We identify specific alterations in two glycine-rich regions as well as aromatic and/or positively charged residues of the IDL by which a broad-range, selective modification of condensation propensity and condensate thermal and chemical stability can be achieved, while leaving the single-stranded DNA and partner protein binding functions of SSB unchanged. AlphaFold 3-predicted structures of tetrameric wild-type and engineered EcSSB constructs identify multiple possible binding sites for the conserved C-terminal tip on the tetramer core of the IDL, establishing a link between condensation propensity and restrictions in IDL conformational dynamics. Besides defining the contributions of IDL-driven interactions to driving protein condensation, these results pave the way for the definition of in vivo roles of EcSSB condensation via genetic engineering and delineate ways for further development of liquid-liquid phase separation prediction algorithms.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures





References
MeSH terms
Substances
Grants and funding
- PD-146123/NKFIH
- VEKOP-2.3.3-15-2016-00007/NKFIH
- PREMIUM-2017-17/Hungarian Academy of Sciences
- ÚNKP-21-3/Ministry for Innovation and Technology
- ÚNKP-19-2/Ministry for Innovation and Technology
- Ministry of Innovation and Technology
- BO/00174/22/Hungarian Academy of Sciences
- BO/00566/24/Hungarian Academy of Sciences
- EKÖP-24-4-II-ELTE-92/Hungarian Academy of Sciences
- EKÖP-24-4-I-ELTE-184/Hungarian Academy of Sciences
- Ministry for Culture and Innovation
- 2018-1.2.1-NKP-2018-00005/Hungarian Ministry for Innovation and Technology
- 2018-1.2.1-NKP/National Research, Development and Innovation Fund
- 101160233/European Union
- HUN-REN Hungarian Research Network
LinkOut - more resources
Full Text Sources

