Validating data from multiplex assays of variant effect: A CanVIG-UK national survey of NHS clinical scientists
- PMID: 40480200
- PMCID: PMC12256791
- DOI: 10.1016/j.ajhg.2025.04.006
Validating data from multiplex assays of variant effect: A CanVIG-UK national survey of NHS clinical scientists
Abstract
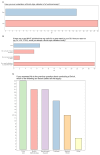
Advances in technology have made it possible for multiplex assays of variant effect (MAVEs) to systematically generate functional data for thousands of genetic variants. Robust clinical validation and accessible online resources for MAVE data have previously been identified as barriers to the clinical adoption of new MAVEs. We delivered a survey during the November 2024 Cancer Variant Interpretation Group UK (CanVIG-UK) meeting comprising National Health Service (NHS) clinical scientists and clinical geneticists and received 46 responses from individuals regularly performing variant classification for diagnostic reporting. Only 35% reported they would accept clinical validation of the MAVE provided by the authors who conducted the assay; 20% reported they would attempt clinical validation themselves, and 61% would await clinical validation by a trusted central body. 72% reported they would use MAVE data ahead of a formal peer-reviewed publication if reviewed and clinically validated by a trusted central body. When scoring central bodies on a scale of 1-5 for confidence in their review and validation of MAVEs, CanVIG-UK (median = 5), variant curation expert panels (VCEPs; median = 5), and ClinGen SVI Functional Working Group (median = 4) all scored highly. Participants supported making variant-level data accessible via a relevant web resource (although the majority of participants expressed that additional assay-level or variant-level information would have a low likelihood of altering validation scores provided by a trusted central body). These findings, from a comparatively homogeneous clinical diagnostic group operating in a resource-constrained healthcare setting, indicate that clinical application of new MAVEs for variant classification will be delayed unless robust clinical validations are performed by a trusted central body and made readily accessible.
Keywords: ACMG/AMP variant classification framework; Brnich-style validation; CanVIG-UK; MAVE data; PS3/BS3 scores; clinical variant classification; variant truth sets.
Copyright © 2025 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Dawood M, Fayer S, Pendyala S, Post M, Kalra D, Patterson K, Venner E, Muffley LA, Fowler DM, Rubin AF, et al. Using multiplexed functional data to reduce variant classification inequities in underrepresented populations. Genome medicine. 2024;16:143. doi: 10.1186/s13073-024-01392-7. - DOI - PMC - PubMed
-
- Chen E, Facio FM, Aradhya KW, Rojahn S, Hatchell KE, Aguilar S, Ouyang K, Saitta S, Hanson-Kwan AK, Capurro NN, et al. Rates and Classification of Variants of Uncertain Significance in Hereditary Disease Genetic Testing. JAMA Netw Open. 2023;6:e2339571. doi: 10.1001/jamanetworkopen.2023.39571. - DOI - PMC - PubMed
-
- Caswell-Jin JL, Gupta T, Hall E, Petrovchich IM, Mills MA, Kingham KE, Koff R, Chun NM, Levonian P, Lebensohn AP, et al. Racial/ethnic differences in multiple-gene sequencing results for hereditary cancer risk. Genetics in medicine : official journal of the American College of Medical Genetics. 2018;20:234–239. - PubMed
-
- Tabet D, Parikh V, Mali P, Roth FP, Claussnitzer M. Scalable Functional Assays for the Interpretation of Human Genetic Variation. Annu Rev Genet. 2022;56:441–465. - PubMed
-
- Fayer S, Horton C, Dines JN, Rubin AF, Richardson ME, McGoldrick K, Hernandez F, Pesaran T, Karam R, Shirts BH, et al. Closing the gap: Systematic integration of multiplexed functional data resolves variants of uncertain significance in BRCA1, TP53, and PTEN. American journal of human genetics. 2021;108:2248–2258. doi: 10.1016/j.ajhg.2021.11.001. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

