Cholangiocytes contribute to hepatocyte regeneration after partial liver injury during growth spurt in zebrafish
- PMID: 40480975
- PMCID: PMC12144294
- DOI: 10.1038/s41467-025-60334-y
Cholangiocytes contribute to hepatocyte regeneration after partial liver injury during growth spurt in zebrafish
Abstract
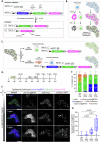
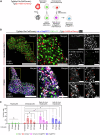
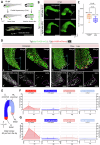
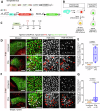
The liver's regenerative ability depends on injury extent. Minor injuries are repaired by hepatocyte self-duplication, while severe damage triggers cholangiocyte involvement in hepatocyte recovery. This paradigm is well-documented for adult animals but is less explored during rapid growth. We design two partial liver injury models in zebrafish, which were investigated during growth spurts: 1) partial ablation, killing half the hepatocytes; and 2) partial hepatectomy, removing half a liver lobe. In both injuries, de novo hepatocytes emerged alongside existing ones. Single-cell transcriptomics and lineage tracing with Cre-driver lines generated by genome editing identified cholangiocytes as the source of de novo hepatocytes. We further identify active mTORC1 signalling in the uninjured liver of growing animal to be a regulator of the enhanced plasticity of cholangiocytes. Our study suggests cholangiocyte-to-hepatocyte transdifferentiation as the primary mechanism of liver regeneration during periods of rapid growth.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
- 40005588/Fonds De La Recherche Scientifique - FNRS (Belgian National Fund for Scientific Research)
- 40013427/Fonds De La Recherche Scientifique - FNRS (Belgian National Fund for Scientific Research)
- 40020360/Fonds De La Recherche Scientifique - FNRS (Belgian National Fund for Scientific Research)
- 40006730/Fonds De La Recherche Scientifique - FNRS (Belgian National Fund for Scientific Research)
- 318346496 - SFB1292/2 TP19N/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

