TANGO2 binds crystallin alpha B and its loss causes desminopathy
- PMID: 40480980
- PMCID: PMC12144310
- DOI: 10.1038/s41467-025-60563-1
TANGO2 binds crystallin alpha B and its loss causes desminopathy
Abstract
Mutations in the TANGO2 gene cause an autosomal recessive disorder characterised by developmental delay, stress-induced episodic rhabdomyolysis, and cardiac arrhythmias along with severe metabolic crises. Although TANGO2 mutations result in a well characterised disease pathology, the function of TANGO2 is still unknown. To investigate the function of TANGO2, we knocked out the TANGO2 gene in human cells and mice. We identify that loss of TANGO2 impairs intermediate filament structure, resulting in fragmented mitochondrial networks and formation of cup-like mitochondria. In male mice, loss of TANGO2 caused heart defects, reduced muscle function and glucose intolerance by remodelling of intermediate filaments, which altered the mitochondrial and cytoplasmic proteomes, N-glycosylation and nucleocytoplasmic O-GlcNAcylation. We identify that TANGO2 binds the small heat shock protein crystallin alpha B (CRYAB) to prevent the aggregation of the intermediate filament desmin and in the absence of TANGO2, mice develop desminopathy, which is consistent with features found in patients carrying mutations in either desmin or CRYAB.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
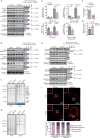



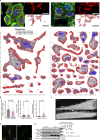
References
-
- Jennions, E. et al. TANGO2 deficiency as a cause of neurodevelopmental delay with indirect effects on mitochondrial energy metabolism. J. Inherit. Metab. Dis.42, 898–908 (2019). - PubMed
-
- Milev, M. P. et al. The phenotype associated with variants in TANGO2 may be explained by a dual role of the protein in ER-to-Golgi transport and at the mitochondria. J. Inherit. Metab. Dis.44, 426–437 (2021). - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

