Dysregulation of cell migration by matrix metalloproteinases in geleophysic dysplasia
- PMID: 40481143
- PMCID: PMC12144124
- DOI: 10.1038/s41598-025-04666-1
Dysregulation of cell migration by matrix metalloproteinases in geleophysic dysplasia
Abstract
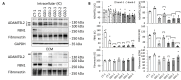
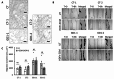
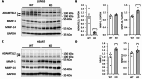
Geleophysic dysplasia (GD) is characterized by short stature, brachydactyly, joint limitations, a distinctive facial appearance, as well as cardiac and respiratory dysfunction that can be life-threatening. GD is caused by pathogenic variants in the ADAMTSL2, FBN1, or LTBP3 genes. While dermal fibroblasts derived from affected individuals have shown poor organization of the extracellular matrix (ECM), it remains elusive how the disorganized ECM contributes to GD pathogenesis. To understand the molecular mechanisms in GD, we isolated and characterized primary human dermal fibroblasts from affected individuals with ADAMTSL2 and FBN1 variants. We found that the secretion of ECM proteins including ADAMTSL2, FBN1, and Fibronectin were impaired in GD fibroblasts. Increased cell migration was observed in GD fibroblasts carrying ADAMTSL2 or FBN1 variants, which was associated with up-regulation of MMP-1 and MMP-14, two proteases related to cell mobility. The enhanced cell migration and up-regulation of MMP-1 and MMP-14 were corroborated in mouse primary dermal fibroblasts carrying pathogenic variants in Adamtsl2 and in lung and heart tissues from Adamtsl2-knockout mice. A pan MMP inhibitor, GM6001, inhibited the migration of GD fibroblasts. Overall, our results suggest that MMP-1/-14 up-regulation play a role in the development of GD and may be utilized as a treatment target.
Keywords: ADAMTSL2; FBN1; Geleophysic dysplasia; MMP-1; MMP-14; MMPs.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Adamtsl2 deletion results in bronchial fibrillin microfibril accumulation and bronchial epithelial dysplasia--a novel mouse model providing insights into geleophysic dysplasia.Dis Model Mech. 2015 May;8(5):487-99. doi: 10.1242/dmm.017046. Epub 2015 Mar 11. Dis Model Mech. 2015. PMID: 25762570 Free PMC article.
-
Limb- and tendon-specific Adamtsl2 deletion identifies a role for ADAMTSL2 in tendon growth in a mouse model for geleophysic dysplasia.Matrix Biol. 2019 Sep;82:38-53. doi: 10.1016/j.matbio.2019.02.001. Epub 2019 Feb 7. Matrix Biol. 2019. PMID: 30738849 Free PMC article.
-
Geleophysic and acromicric dysplasias: natural history, genotype-phenotype correlations, and management guidelines from 38 cases.Genet Med. 2021 Feb;23(2):331-340. doi: 10.1038/s41436-020-00994-x. Epub 2020 Oct 21. Genet Med. 2021. PMID: 33082559
-
A report of three families with FBN1-related acromelic dysplasias and review of literature for genotype-phenotype correlation in geleophysic dysplasia.Eur J Med Genet. 2018 Apr;61(4):219-224. doi: 10.1016/j.ejmg.2017.11.018. Epub 2017 Nov 27. Eur J Med Genet. 2018. PMID: 29191498 Review.
-
From tall to short: the role of TGFβ signaling in growth and its disorders.Am J Med Genet C Semin Med Genet. 2012 Aug 15;160C(3):145-53. doi: 10.1002/ajmg.c.31337. Epub 2012 Jul 12. Am J Med Genet C Semin Med Genet. 2012. PMID: 22791552 Review.
Cited by
-
Case Report: A case of severe pulmonary hypertension combined with FBN1 mutation associated geleophysic dysplasia.Front Pediatr. 2025 Jul 16;13:1642390. doi: 10.3389/fped.2025.1642390. eCollection 2025. Front Pediatr. 2025. PMID: 40740820 Free PMC article.
References
-
- Marzin, P. & Cormier-Daire, V. Geleophysic Dysplasia. In GeneReviews® [Internet] (eds Adam, M. P. et al.) (University of Washington, Seattle) (1993–2024). - PubMed
-
- Marzin, P. et al. Geleophysic and acromicric dysplasias: natural history, genotype-phenotype correlations, and management guidelines from 38 cases. Genet. Medicine: Official J. Am. Coll. Med. Genet.23, 331–340 (2021). - PubMed
-
- Spranger, J. W., Gilbert, E. F., Tuffli, G. A., Rossiter, F. P. & Opitz, J. M. Geleophysic dwarfism–a focal. mucopolysaccharidosis? Lancet (London England). 2, 97–98 (1971). - PubMed
-
- Spranger, J., Gilbert, E. F., Arya, S., Hoganson, G. M. I. & Opitz, J. M. Geleophysic Dysplasia19, 487–499 (1984). - PubMed
-
- Lipson, A. H., Kan, A. E. & Kozlowski, K. Geleophysic dysplasia-acromicric dysplasia with evidence of glycoprotein storage. Am. J. Med. Genet. Suppl.3, 181–189 (1987). - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

