crossNN is an explainable framework for cross-platform DNA methylation-based classification of tumors
- PMID: 40481322
- PMCID: PMC12296554
- DOI: 10.1038/s43018-025-00976-5
crossNN is an explainable framework for cross-platform DNA methylation-based classification of tumors
Abstract
DNA methylation-based classification of (brain) tumors has emerged as a powerful and indispensable diagnostic technique. Initial implementations used methylation microarrays for data generation, while most current classifiers rely on a fixed methylation feature space. This makes them incompatible with other platforms, especially different flavors of DNA sequencing. Here, we describe crossNN, a neural network-based machine learning framework that can accurately classify tumors using sparse methylomes obtained on different platforms and with different epigenome coverage and sequencing depth. It outperforms other deep and conventional machine learning models regarding accuracy and computational requirements while still being explainable. We use crossNN to train a pan-cancer classifier that can discriminate more than 170 tumor types across all organ sites. Validation in more than 5,000 tumors profiled on different platforms, including nanopore and targeted bisulfite sequencing, demonstrates its robustness and scalability with 99.1% and 97.8% precision for the brain tumor and pan-cancer models, respectively.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.C. is a shareholder and cofounder of Heidelberg Epignostix. All other authors declare no competing interests.
Figures





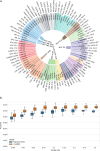
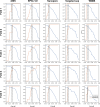

References
-
- Klutstein, M., Nejman, D., Greenfield, R. & Cedar, H. DNA methylation in cancer and aging. Cancer Res.76, 3446–3450 (2016). - PubMed
-
- Nishiyama, A. & Nakanishi, M. Navigating the DNA methylation landscape of cancer. Trends Genet.37, 1012–1027 (2021). - PubMed
-
- Papanicolau-Sengos, A. & Aldape, K. DNA methylation profiling: an emerging paradigm for cancer diagnosis. Annu. Rev. Pathol.17, 295–321 (2022). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

