Modular and signal-responsive transcriptional regulation using CRISPRi-aided genetic switches in Escherichia coli
- PMID: 40481524
- PMCID: PMC12142916
- DOI: 10.1186/s13036-025-00526-8
Modular and signal-responsive transcriptional regulation using CRISPRi-aided genetic switches in Escherichia coli
Abstract
Background: Precise and dynamic transcriptional regulation is a cornerstone of synthetic biology, enabling the construction of robust genetic circuits and programmable cellular systems. However, existing regulatory tools are often limited by issues such as leaky transcription and insufficient tunability, particularly in high-expression or complex genetic contexts. This study aimed to develop a CRISPRi-aided genetic switch platform that overcomes these limitations and expands the functionality of transcriptional regulation tools in synthetic biology.
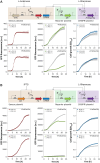
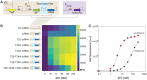
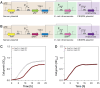
Results: We established a versatile CRISPRi-aided genetic switch platform by integrating transcription factor-based biosensors with the Type V-A FnCas12a CRISPR system. Exploiting the RNase activity of FndCas12a, this system processes CRISPR RNAs (crRNAs) directly from biosensor-responsive mRNA transcripts, enabling precise, signal-dependent transcriptional regulation. To mitigate basal transcription and enhance regulatory precision, transcriptional terminator filters were incorporated, reducing leaky expression and increasing the dynamic range of target gene regulation. The platform demonstrated exceptional adaptability across diverse applications, including ligand-inducible genetic switches for transcriptional control, signal amplification circuits for enhanced output, and metabolic genetic switches for pathway reprogramming. Notably, the metabolic genetic switch dynamically repressed the endogenous gapA gene while compensating with orthologous gapC expression, effectively redirecting metabolic flux to balance cell growth.
Conclusions: The CRISPRi-aided genetic switch provides a powerful and flexible toolkit for synthetic biology, addressing the limitations of existing systems. By enabling precise and tunable transcriptional regulation, it offers robust solutions for a wide array of biotechnological applications, including pathway engineering and synthetic gene networks.
Keywords: CRISPR interference; Genetic switch; Metabolic genetic switch; Signal amplifier; Transcriptional regulation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
CRISPR interference-guided multiplex repression of endogenous competing pathway genes for redirecting metabolic flux in Escherichia coli.Microb Cell Fact. 2017 Nov 3;16(1):188. doi: 10.1186/s12934-017-0802-x. Microb Cell Fact. 2017. PMID: 29100516 Free PMC article.
-
CRISPRi/dCpf1-mediated dynamic metabolic switch to enhance butenoic acid production in Escherichia coli.Appl Microbiol Biotechnol. 2020 Jun;104(12):5385-5393. doi: 10.1007/s00253-020-10610-2. Epub 2020 Apr 27. Appl Microbiol Biotechnol. 2020. PMID: 32338294
-
Overcoming Leak Sensitivity in CRISPRi Circuits Using Antisense RNA Sequestration and Regulatory Feedback.ACS Synth Biol. 2022 Sep 16;11(9):2927-2937. doi: 10.1021/acssynbio.2c00155. Epub 2022 Aug 26. ACS Synth Biol. 2022. PMID: 36017994 Free PMC article.
-
CRISPR-Based Approaches for Gene Regulation in Non-Model Bacteria.Front Genome Ed. 2022 Jun 23;4:892304. doi: 10.3389/fgeed.2022.892304. eCollection 2022. Front Genome Ed. 2022. PMID: 35813973 Free PMC article. Review.
-
Prospects for engineering dynamic CRISPR-Cas transcriptional circuits to improve bioproduction.J Ind Microbiol Biotechnol. 2018 Jul;45(7):481-490. doi: 10.1007/s10295-018-2039-z. Epub 2018 May 8. J Ind Microbiol Biotechnol. 2018. PMID: 29740742 Review.
References
-
- Cashin P, et al. Contrasting signal transduction mechanisms in bacterial and eukaryotic gene transcription. FEMS Microbiol Lett. 2006;261(2):155–64. - PubMed
-
- Bourret RB, Borkovich KA, Simon MI. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–41. - PubMed
-
- Hojfeldt JW, Van Dyke AR, Mapp AK. Transforming ligands into transcriptional regulators: Building blocks for bifunctional molecules. Chem Soc Rev. 2011;40(8):4286–94. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

