Integrative multi-omics analysis reveals the LncRNA 60967.1-PLCD4-ATRA axis as a key regulator of colorectal cancer progression and immune response
- PMID: 40481569
- PMCID: PMC12142938
- DOI: 10.1186/s12943-025-02359-x
Integrative multi-omics analysis reveals the LncRNA 60967.1-PLCD4-ATRA axis as a key regulator of colorectal cancer progression and immune response
Abstract
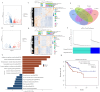
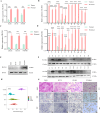
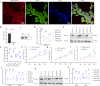
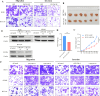
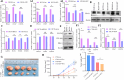
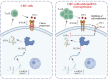
Colorectal cancer (CRC) is a major global health concern, characterized by high morbidity and mortality rates. CRC progression involves intricate molecular networks that remain incompletely understood. In this study, we conducted an integrative multi-omics analysis of transcriptomic, proteomic, and metabolomic profiles from CRC tissues and matched normal adjacent tissues (NATs). Our analysis revealed 1,394 differentially expressed long non-Coding RNAs (lncRNAs), 2,788 genes, 548 proteins, and 91 metabolites. A significant interaction network comprising 22 lncRNAs, 14 mRNAs/proteins, and 9 metabolites was identified, among which lncRNA 60967.1 emerged as a pivotal regulator. Functional validation demonstrated that lncRNA 60967.1 is markedly downregulated in CRC cell lines and patient tissues. Overexpression of lncRNA 60967.1 restored expression of the tumor suppressor PLCD4 and increased levels of all-trans retinoic acid (ATRA). This modulation enhanced IFN-γ-induced apoptosis and increased expression of the IFN-γ receptor subunit IFNGR1, thereby partially reversing IFN-γ resistance. In murine models, lncRNA 60967.1 overexpression promoted immune cell infiltration and synergized with anti-PD-1 therapy to inhibit tumor growth. Collectively, our findings uncover a novel lncRNA-mRNA/protein-metabolite network, the lncRNA 60967.1-PLCD4-ATRA axis, that plays a critical role in CRC progression and immune modulation, offering promising therapeutic targets for improved treatment efficacy.
Keywords: Colorectal cancer; Immunity; LncRNAs; Multi-omics; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study received ethical approval from Sir Run Run Shaw Hospital, Hangzhou, China. All patients provided written informed consent. The Institutional Animal Care and Use Committee (IACUC) of the respective institutions approved all experiments. Consent for publication: All the authors have read and approved the final manuscript for publication. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Comprehensive analysis of the long noncoding RNA expression profile and construction of the lncRNA-mRNA co-expression network in colorectal cancer.Cancer Biol Ther. 2020;21(2):157-169. doi: 10.1080/15384047.2019.1673098. Epub 2019 Oct 16. Cancer Biol Ther. 2020. PMID: 31619123 Free PMC article.
-
Long non-coding RNA RP11-197K6.1 as ceRNA promotes colorectal cancer progression via miR-135a-5p/DLX5 axis.J Transl Med. 2024 May 17;22(1):469. doi: 10.1186/s12967-024-05286-5. J Transl Med. 2024. PMID: 38760791 Free PMC article.
-
Transcriptome profiling and co-expression network analysis of lncRNAs and mRNAs in colorectal cancer by RNA sequencing.BMC Cancer. 2022 Jul 16;22(1):780. doi: 10.1186/s12885-022-09878-6. BMC Cancer. 2022. PMID: 35842644 Free PMC article.
-
Comprehensive analysis of long noncoding RNA and mRNA in five colorectal cancer tissues and five normal tissues.Biosci Rep. 2020 Feb 28;40(2):BSR20191139. doi: 10.1042/BSR20191139. Biosci Rep. 2020. PMID: 32016349 Free PMC article.
-
Identification of MFI2-AS1, a Novel Pivotal lncRNA for Prognosis of Stage III/IV Colorectal Cancer.Dig Dis Sci. 2020 Dec;65(12):3538-3550. doi: 10.1007/s10620-020-06064-1. Epub 2020 Jan 20. Dig Dis Sci. 2020. PMID: 31960204 Review.
References
-
- Morgan E, Arnold M, Gini A, Lorenzoni V, Cabasag CJ, Laversanne M, Vignat J, Ferlay J, Murphy N, Bray F. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut. 2023;72(2):338–44. - PubMed
-
- Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73(3):233–54. - PubMed
-
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–64. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

