Detection of KPC-producing Enterobacterales species in wastewater samples from the Gran Concepción Metropolitan area, Chile
- PMID: 40481605
- PMCID: PMC12144836
- DOI: 10.1186/s40659-025-00612-7
Detection of KPC-producing Enterobacterales species in wastewater samples from the Gran Concepción Metropolitan area, Chile
Abstract
Background: Carbapenemase-mediated resistance to carbapenems is a significant public health concern due to its potential for widespread dissemination. The KPC family of carbapenemases, encoded by the blaKPC gene and often associated with Tn4401-like transposons, is particularly important for its ability to be transferred through diverse plasmid types. In Chile, KPC-producing Gram-negative bacteria have been detected in clinical settings; however, their occurrence in wastewater (WW) remains unknown. This study addresses this gap by characterizing carbapenem-resistant Enterobacterales isolates from a wastewater treatment plant (WWTP) in the Gran Concepción Metropolitan Area, Chile.
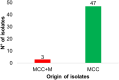
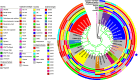
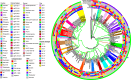
Results: This study identifies three carbapenem-resistant Enterobacterales isolates, namely Klebsiella pasteurii M2/A/C/34, Klebsiella pneumoniae subsp. pneumoniae M3/A/M/3, and Citrobacter freundii sensu stricto. M4/A/C/32, all exhibiting multidrug-resistant profiles and carrying the blaKPC-2 gene encoding KPC-like carbapenemases. These isolates also possessed genes for extended-spectrum β-lactamases (ESBLs) and aminoglycoside-modifying enzymes (AMEs). Sequence typing revealed that M2/A/C/34, M3/A/M/3, and M4/A/C/32 belonged to novel sequence types, specifically ST470, ST273, and ST214, respectively. All isolates carried plasmids belonging to groups commonly associated with ARGs, including IncF, IncP, and IncA. Both Klebsiella isolates (M2/A/C/34 and M3/A/M/3) carried the class 1 integron (intl1) gene. Phylogenomic analysis reveals that M2/A/C/34 is related to strains from China and Pakistan, while M3/A/M/3 shares similarities with a strain from Germany, indicating their potential dissemination.
Conclusions: This study represents the first detection of carbapenem-resistant Enterobacterales carrying blaKPC-2 in Chilean WW, including the novel identification of K. pasteurii. These findings emphasize the critical role of genomic surveillance in WW under the One Health framework, enabling the monitoring of carbapenemase-producing bacteria and associated ARGs. Sustained surveillance efforts are essential to comprehend the dynamics of antibiotic resistance in environmental reservoirs and to develop strategies for its containment and mitigation.
Keywords: Antibiotic resistance; Carbapenem-resistant Enterobacterales; KPC-type carbapenemases; One health; Wastewater.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- WHO. Bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance: World Health Organization; 2024. https://www.who.int/publications/i/item/9789240093461 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

